Last week we announced the winners of the ASAPbio competition ‘Make your negative result a preprint winner’, a project by a group of ASAPbio Fellows that aimed to celebrate the value of using preprints to share negative and inconclusive scientific results. We have talked to the authors of the winning preprints to learn more about their work and their motivation to post their results as a preprint. In this post, Mark Hanson (École Polytechnique Fédérale de Lausanne), the first author of the preprint ‘Antimicrobial peptides do not directly contribute to aging in Drosophila, but improve lifespan by preventing dysbiosis‘, tells us about the journey to collect the results in this project and why he and his co-author felt it was important to share these negatives results with the community.
Tell us a little bit about the research project and results reported in the preprint
The research project was initially part of a larger study to finally characterize how these genes called “antimicrobial peptides” or “AMPs” work. These are short immune effectors, basically host-encoded antimicrobials that defend you against infection. One of the ways they do this is through their positive charge, which disrupts the negative charge of microbe membranes. During aging, it’s very common to see activation of immunity (so-called “inflammaging”), and other inflammatory syndromes (e.g. inflammatory bowel disease) are in close association with microbes and host immunity. It’s been a longstanding question whether AMPs, produced as part of the immune response, might be negatively contributing to aging and other inflammatory syndromes. Host cell membranes are usually neutral in charge. However, a few studies using genetic tools in model systems, saw damage to host cells when they overexpressed AMPs, or reduced damage in neurodegenerative disease models. There are some caveats to using such genetic tools… but they are typically the best approach we have to delve deeper; and they are quite powerful for the right questions! The idea generated by these studies was that aging might be exacerbated by AMPs because as cells get older and deal worse with stress, they might change their properties to become sensitive to AMPs; for instance, host cells can shift the charge of their cell membranes to become more negative during stress, which might make them vulnerable to collateral damage from positively-charged AMPs produced during inflammatory responses. So we were testing this “AMPs damage the host” model.
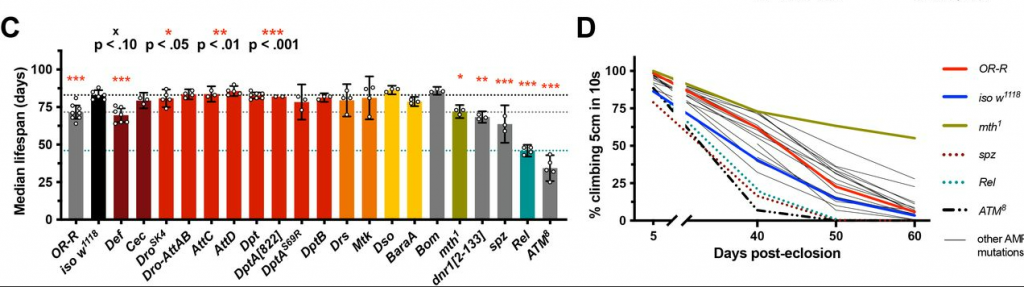
Our lab generated the first systematic loss-of-function mutations for AMPs in an animal model: Drosophila fruit flies. Using those special flies, we could ask what each AMP gene was doing individually, or in systematic combinations where we deleted one, two, three… all the way up to 14 genes simultaneously. We then checked the aging of those flies to see if deleting AMPs helped them live longer, which is what you’d predict given the “AMPs damage the host” model. But if anything we saw the opposite: flies lacking individual AMPs were fine, so it’s not like any one gene was making a difference. And flies lacking multiple genes never lived better than normal flies, while certain combinations actually did worse (including flies lacking 14 AMP genes simultaneously).
We re-tried the experiments in flies reared in sterile conditions deprived of their bacteria. Our thinking was: “Ok, even if AMPs do damage the host, but they also are obviously important for keeping microbes under control. If we get rid of the microbes, maybe we’ll see the AMP mutants live longer than wild-type flies.” But when we did that, we didn’t even rescue the AMP mutants up to the lifespans of their wild-type controls. We DID improve their lifespan by quite a bit! So in the absence of AMPs, microbes grow out of control and that’s harmful. But we didn’t find any evidence for this longstanding “AMPs damage the host” model. We show that, under normal conditions, if AMPs have an intrinsic effect on lifespan by damaging the host, it’s probably fairly minor in the grand scheme of things. They could still be specifically important in certain disease contexts, but it seems unlikely that AMP responses are a major factor that accelerates aging more generally.
What would you say is the most valuable contribution of your results to your community?
There are two answers to this question: 1) I think our results tamp down a bit of the excitement around AMPs and aging. Again, that’s not to say they can’t be playing neat roles in aging and neurodegenerative diseases! I just saw a cool paper that found the AMP Metchnikowin might be contributing to a very specific type of neurodegeneration model, using the same mutants we produced as part of our study; we also saw a slight improvement in climbing ability of Metchnikowin mutants, and another study saw that deleting Metchnikowin improved fly responses to traumatic brain injury. So there’s probably something there! But as far as your standard life process goes? Our results would suggest that Metchnikowin, or indeed most other AMPs, aren’t major players. That should hopefully focus the field and better think about what it means when you see these AMPs being expressed during aging. Are they cause, or effect? Our study would argue they’re more effect, less cause. 2) We also noted this cryptic viral infection that confused our results for like… 2.5 years. We decided to make this our Figure 1, front and centre, so we could hopefully bring awareness of this infection to the field. In our hands, with our fly strains, it had a much larger effect than what a previous study reported. So we felt it was important to get the word out there.
Why did the group decide to post a preprint with these results? Was this an easy decision for all the co-authors?
We’ve been preprinting all our manuscripts for years now, so it was an easy decision. I don’t see why we wouldn’t? It makes the study, in a mature form, freely available to the public no matter what. It might change following review, but that can be updated on preprint servers. Indeed, we changed the way the data was presented following review, but the key results remained the same, and unless you were paying close attention, you probably wouldn’t even realize there were any changes to the figures or underlying data. So preprints really are communicating the most important parts of the science as soon as they’re ready, with the bonus of being able to refine that study for final publication later.
Did you submit the paper for publication in a journal? If not, why not?
Yes we did. In fact, it was just accepted for publication and put online in pre-publication form! We used Review Commons to get peer reviews, and then took our manuscript with the reviews in hand to the journal we thought was the best fit given the reviewer enthusiasm and the comments/critiques. It was accepted in Disease Models and Mechanisms following some back-and-forth between us, one reviewer in particular, and the editor. Thank you editor for being so patient during this process! And thank you to the reviewer for the constructive feedback, even if we didn’t see eye-to-eye on everything.
Would you encourage other researchers to write up negative results as preprints? How can we better recognize the value of negative results?
Absolutely. In our case, this wasn’t an “exciting” project in the sense that nothing seemed to go in the direction we expected, and our data collection was complicated by this cryptic infection… it was 5 years of work that didn’t really advance the current model at all! But we learned a number of lessons from doing that work:
i) we observed this more drastic importance of cryptic virus infection than previous studies would suggest.
ii) we collected a lot of data on the lifespans of many fly strains that people were just beginning to use for many different kinds of questions, so we might as well make them public.
iii) despite a number of experiments (including stuff we still haven’t published, and likely never will), we realized that we weren’t getting anywhere with the “AMPs damage the host” model that motivated our research approach. By publishing our results, hopefully we can avoid someone else going through the same workload to find out what we already knew – or at least provide some context if a future study ends up disagreeing!
iv) because we didn’t really find the result we were expecting, we made a point of taking a step back and asking why it was that the field had this model of aging in the first place. That motivated us to put a lot of control genotypes into our study for the sake of measuring lifespan in control flies that weren’t part of our research question. It was a way we could do an internal “sanity check” on our procedures, and hopefully make it easier for others to compare their results to our own. I personally hope that more aging studies take that approach on board, because there can be such huge “butterfly effects” when you’re measuring a process that’s months – or years-long… and where the strains/genetic tools you use all come with caveats, and then of course the conditions in research groups can differ in imperceptible ways… it was useful for us to sort through the noise in the literature (and our own data) for what was likely real and a major effect, and what might be real, or might just be noise in the data.
Next week, we’ll share our conversation with Lilya Andrianova about the research behind the winning preprint ‘No evidence from complementary data sources of a direct projection from the mouse anterior cingulate cortex to the hippocampal formation’, stay tuned!







