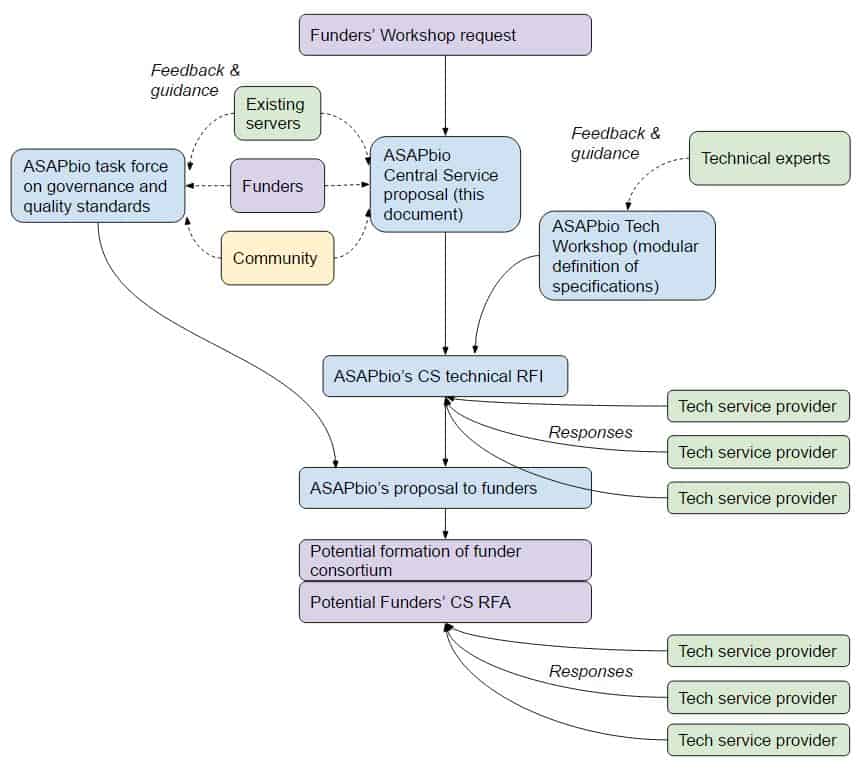
Central Service model documents
- Summary: Background and a draft model
- Appendix 1: Rationale for a Central Service
- Appendix 2: Current feedback on Central Service features
Current discussions with the community on proposed features of the Central Service
Surveys and information from scientists
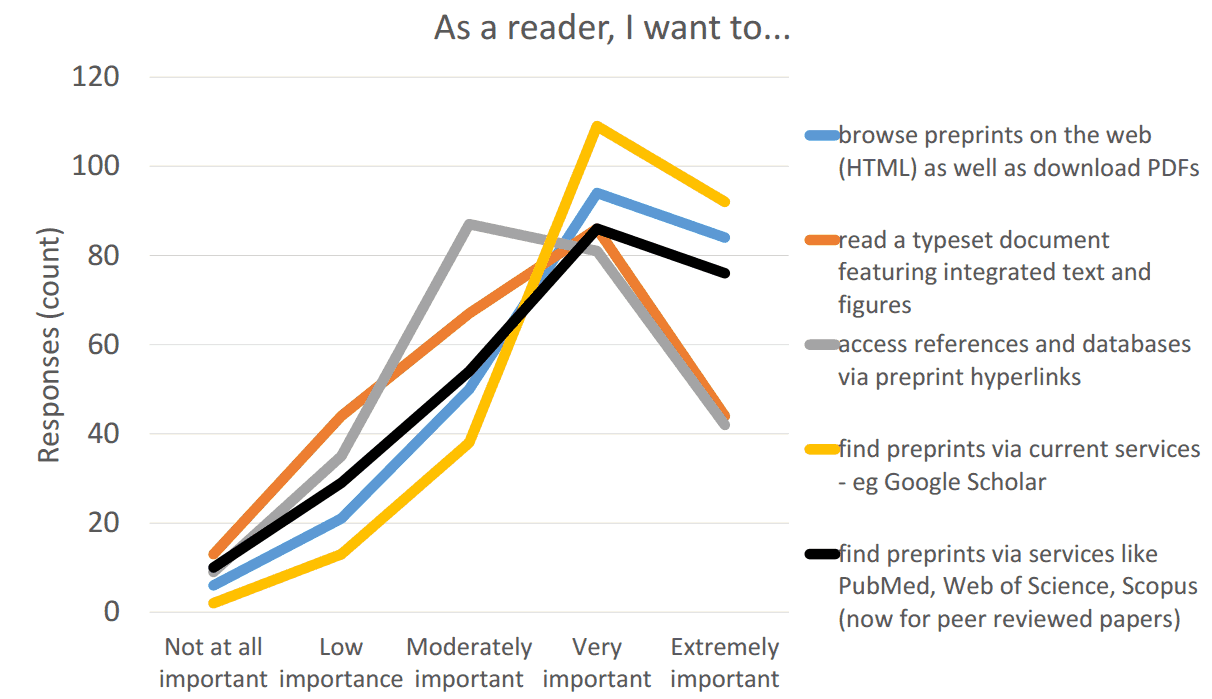
We will continue to engage the scientific community on what services they want to see in a next generation of preprints. However, based upon a survey that ASAPbio conducted in May 2016 (Results summary (pdf) and Anonymized responses (xls)) and other resources (e.g. Preprint user stories compiled by Jennifer Lin at Crossref and ASAPbio survey #1 (early 2016)), we believe that biologists want:
- High visibility and discoverability of preprints
- A single recognized website
- Good search tool
- Email notifications
- Web-readable xml format
- Click on link to figures to display them
- Ability to click on links to references
- Export to more readable and compact pdfs
- A system for cross-referencing versions of the same work
- Linking the final journal publication to preprint versions (and vice versa), so that the history of the work is transparent and preserved
Input from servers, publishers, funders and data management experts
In July-August 2016, ASAPbio conducted informal interviews with preprint servers, funders, scientists and developers. We originally presented a variety of Central Preprint Service models of increasing complexity and centralization, ranging from a PubMed-like metadata search tool (Model 1) to a PubMed Central-like database that hosts well-formed XML content (JATS) and makes it available through a web display tool and an API (Model 4). One version also included a central submission tool (Model 5).
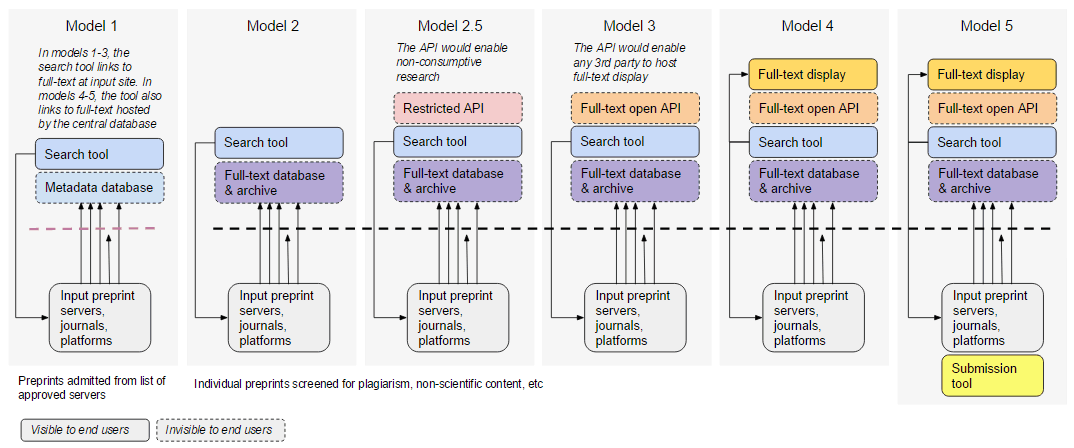
While responses to the creation of a central tool were generally very positive, opinions on the best implementation varied. Below is a summary of some of the critical feedback we received.
- Models 1 & 2 provide little benefit over the current state of affairs. These models generated less interest among funding agencies.There are already multiple ways to search preprints (search.bioPreprint, PrePubMed, Google Scholar) and existing preprint servers already preserve their own content.
- Models without an open API and common licensing will stifle innovation. Without free access to content, 3rd parties will have difficulty in implementing new services (such as peer review, data mining, or aggregation)
- Providing central submission and full-text display would be undesirable for some existing servers. These tools would directly compete with existing servers for traffic and recognition in the community. Also, display in multiple locations could disrupt download/view metrics and commenting systems. However, some funders felt that the CS should have the ability for full display as well as drive traffic to server sites. Some funders have expressed an interest in allowing submission directly to the CS (Model 5), but most favor a practical solution that embraces the needs of the ecosystem.
- Many of the original models are complicated and development of any system with many moving parts will take a long time. Therefore, “perfection must not be the enemy of the good.” There will be a need to generate a CS that will work “out of the box” and improve on it over time. The CS needs to take into account realistic development of technologies.
- Technological limitations make the use of JATS impractical. No good unsupervised .doc -> JATS converters currently exist. Thus, the conversion process requires human intervention.
- Document conversion is costly. Server-side conversion to a structured format (such as JATS) is expensive (on the order of ~$20+); therefore, it doesn’t make sense for preprint servers to provide this, especially when preprints generate no revenue. The CS should be close to cost-neutral to servers and other publishing entities.
- Licensing has generated a diversity of opinion. Some parties favor author or publisher choice in licensing, arguing that scientists will have concerns of the re-use of their material. Our own surveys and interactions with scientists suggest that most do not understand licensing options and their associated benefits/disadvantages. Most funders favor a uniform licensing policy for the material in the CS in order to allow re-use in innovative ways and avoid complicated restrictions for data mining. The license most favored at the moment is CC-BY, although this may require research and engagement with the scientific community.
- Servers, Platforms, Publishers consulted were generally interested in working with the CS. However, alignment and preference for models varied between model 2 and model 4.
- A major topic in which opinion varies is ‘display’. Some funders and publishers the CS should have capability of displaying its archived content. Others feel that the CS should not display content to readers (other than abstracts) and that display should reside with servers and publishers.
- Use existing technologies whenever possible. Don’t reinvent the wheel, and carefully evaluate existing software/infrastructure.
- Balances immediate concerns against opportunities for future development. Expressed by many, this sentiment emphasizes the need for a governance body that can continuously weigh these issues over time and make adjustments. In addition, inter-operability between preprints systems in biology, physics and other disciplines may need to be considered in the future.
We have drafted a provisional model of the Central Service (Summary) that takes into account the various input received above. The model emphasizes the development of document conversion services and the provision of web-ready full-text outputs to the input server. Providing full-text display via intake server/publishers will deliver to scientists many of the benefits they want while providing intake servers with incentives to participate in the program.
Possible benefits of the proposed service
To scientists
- Preservation
- Ease of use and readability (through web display at intake server)
- Adherence to standards of author identity and ethical guidelines for research and disclosure
- Potential for innovative reuse (with appropriate attribution)
To intake servers/platforms
- No-cost document conversion into web-readable format
- No-cost preservation
- Improved exposure through a search portal that links exclusively to the intake server for display
- “Accreditation” of servers or individual preprints through central screening process
To funders
- Uniform standards of quality
- Access to entire corpus via API
- Ability to search/filter by funding source
Desired technical features for discussion
We welcome comments on the list of desired features below, which could become an agenda for discussion at the Technical Workshop (August 30, 2016).
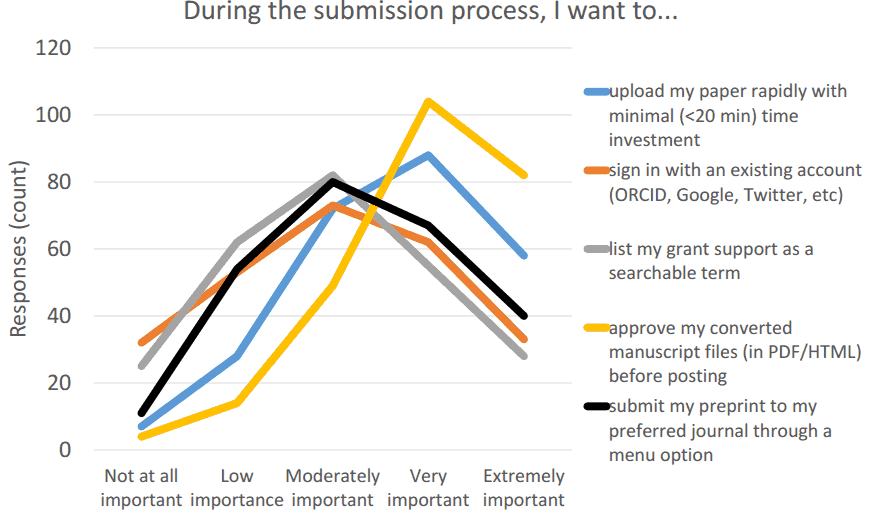
Input (collected from the author)
- Original manuscript file (.doc so that reference metadata can be extracted)
- Supplementary files
- ORCID
- License (note- the Governance Body task force will also address this issue)
- User authentication
- Metadata (if extracted from .doc, get the user to check)
- Grant support
- Ethical statements (note- a separate task will also address this issue)
- Self-ID COI
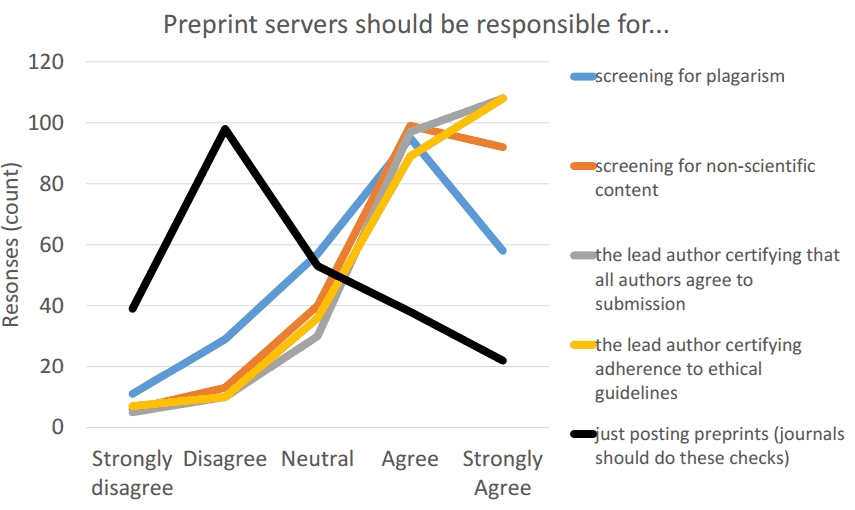
- All authors agree on submission
- Methods needed to reproduce this work are contained within the work
- The work has been conducted in agreement with human & animal research guidelines
Document conversion
- Extraction of text from source file
- Extraction of metadata (such as title, authors, affiliations, keywords, and abstract)
- Extraction of references
- Insertion of figures, or recognition of existing in-line figures
Screening and moderation
- Automated plagiarism detection
- Automated detection of non-scientific content (via arXiv-like algorithm)
- Interface for human-supervised screening/curation/moderation
Versions and identifiers
- Unique, persistent ID for each version
- All versions linked to one another (and to published journal article)
- Linked to datasets
- Tombstone pages for retracted content
Archiving
- Stable archiving of source file (.doc) and also derivatives
- Permission to display content if intake server reaches end of life
API
- Bulk download of all content (.doc) and also derivatives
- Filtering by metadata
Discovery tool
- Full-text indexing of all content in the central database
- Advanced search (boolean operators, search fields such as author, keyword, funding support)
- Alerts (RSS/email)
- Display of abstracts, etc, but exclusive link to intake server for full-text display
Proposal development process
The output of the Technical Workshop will be an announced in a Request For Information (RFI), in response to which any interested party can provide information on the development and approximate costs of developing a CS. The responses to the RFI will be shared by ASAPbio with major international funding agencies for potential consortium support. Pending their collective interest in financially supporting a plan for a CS and refining its method of operation and governance, a formal RFA may follow the RFI to which interested parties could apply for funding.