Your comments welcome
We would like to hear your feedback and reactions, and we especially encourage readers to annotate specific parts of the report with links to existing technologies or projects that may fulfill some of these needs.
We’re using Hypothesis as a tool for social annotation, and you’ll be prompted to register for an account if you don’t already have one.
About this report
In 2020, the Chan Zuckerberg Initiative Open Science Program started a collaboration with ASAPbio to identify shared technology needs for preprints. Preprints have come of age in many areas of science, particularly in the life sciences. The pandemic has brought additional momentum to community innovations that build on preprints. Countless experiments have emerged to unlock the potential of preprints by supporting open collaboration, community curation, and content enrichment, thereby making the overall process of early dissemination of results more transparent and open. That being said, while many of these experiments focused on identifying incentives and strategies to drive participation, they often lacked adequate technical support. This project was set out to identify missing technology that would enhance preprint-based collaboration and innovation. We collected information and user stories (excerpted in each section) from various stakeholders through interviews, surveys, and a workshop. This report summarizes key technology-related preprint needs that emerged from this work. For more information on the methodology and groups who participated in this effort, see the Appendix. For more background on the motivation behind this project, see this piece.
In each section describing an identified need below, user stories emerged from consultations described in the Appendix while desired features and challenges & concerns reflect points raised during the workshop, including those voiced by single participants or challenged by others.
Last updates: 2021-05-17 (Acknowledgements), 2021-05-21 (About this report)
Table of contents
Identified needs
- Markup language conversion
- Better linking from preprints to their journal version
- Better preprint recommenders
- Summaries and badges for review content
- Improved support for preprint review
- Preprint server integration with review platforms
- Standardized review metadata
- Author options for receiving reviews
- Notifications and updates
- Other needs
1. Markup language conversion
Motivation
Many preprint servers host and display only author-submitted PDF files. However, HTML or XML full text preprints provide a better reading experience, including more flexible, responsive, accessible, and mobile-friendly display formats, which are especially important for low- and lower-middle-income countries., Structured full text also makes reference, licenses, and data from figures and tables more accessible, improves full-text search and discovery of preprints, and aids in content preservation. Finally, HTML and XML preprints better support the accurate functioning of automated tools that rely on text and data mining as well as other future innovations (in commenting, links to other resources, reuse of preprints by other tools).
Currently, some servers pay for semi-automated XML conversion by vendors, but this takes time (delaying access to the structured full text, though PDFs can be available immediately) and importantly, financial resources which are inaccessible to many servers, especially those with a large backlog of content. A fully-automated conversion process would be cheaper and faster; it would also make submission easier for authors since it would be possible to extract metadata during submission whilst allowing authors to focus on checking metadata rather than entering it manually.
Example user stories
- As a reader, I want to view articles in different layouts (figures on the side, etc), so that I can more easily cross-reference figures and results. –Evolutionary genetics researcher, Research Institution, India
- As a (student) reader, I want to download a pdf of a preprint in a compact, easy-to-read format, so that I can have a smoother reading experience and stay within my university printing quota. –Cell biologist, Primarily Undergraduate Institution, USA
- As a reader, I want to download all preprint figures as a powerpoint slide so that I can more easily incorporate them into journal clubs and other presentations. –Synthetic biologist, Research Institution & Community lab, USA
Desired features
- Handle formulas and tables properly
- Integrate with a WYSIWYG XML-native editor, enabling authors to:
- Fix problems at submission
- Modify an existing preprint in the tool to create a new updated version
- Compose the manuscript in the editor
- Receive suggested formatting as they write
- Connect to the editor to Microsoft Word to better structure their document
- Agreement on a standardized (non-display) format. E.g. JATS XML (perhaps with more implementation guidance) would simplify the process of conversion to other display formats (HTML, ePUB, DAISY, etc.)
- Agreement on a standardized structured packaging standard (for example MECA) to combine the XML-converted full text with required assets (for example, image files) and, possibly, the original source files
- Integrate comments, badges, annotations, and other innovations with structured full text
- Support legacy formats (e.g., PDFs are still necessary for some screening tools)
- Easy to use
- Accessible to many different preprint servers and platforms
Challenges and concerns
- Some platforms report low usage of full-text HTML. However, this is heavily influenced by the layout and design choices, as other platforms see more use of HTML than PDF.
- Some authors may not want automated conversion because it facilitates for subsequent uses of their documents they may not want.
- A submission process that includes conversion and extensive checking and proofing may become cumbersome, taking away some of the joy and ease of posting preprints.
- Conversion at the preprint stage may duplicate effort by publishers. However, preprint XML could also be reused by publishers if workflows permitted this.
- Professional-looking preprints could be mistaken for peer reviewed articles, or shift focus away from them.
2. Better linking from preprints to their journal version
Motivation
As manuscripts that typically have not been formally peer reviewed may undergo further revisions, and are often cited as preliminary findings, preprints need a robust linking solution to connect them to their corresponding journal article. These links will ensure readers can see a paper’s most recent version and will provide readers with a transparent record of the evolution of a paper, including when a preprint is cited in another paper.
Example user stories
- As a reader, I want to see links on preprints to their journal version, so that I can keep up to date with research. –Evolutionary genetics researcher, Research Institution, India
- As an author, I want to have robust, automatic linking from my preprint to the published journal article, so that I can help readers discover the updated version of the paper. –Bioengineering researcher, Minority Serving Institution, USA
- As a reader, I want to see a link to the journal version from the preprint (once published), so that I can keep up with any updates. –Synthetic biologist, Community lab, USA
Desired features
- Allow authors and third parties to assert preprint links (for example, when authors add preprints to their ORCiD profile)
- Filter preprint/journal links by provenance (server or journal, author, or third party)
- Feedback mechanism to request metadata fixes
- Include links to corrections, retractions, or other notices
- Enable preprint servers to update metadata automatically
- Notify preprint servers if an article is retracted; notify publishers if a preprint is withdrawn or removed
- Keep all versions of metadata changes (e.g. when a new preprint version is posted or updated with a link to the journal versions) and make logs of changes to metadata widely accessible
Challenges and concerns
- Publishers want to maximize traffic to the journal article and may not add links back to the preprint.
- If a link to a journal article becomes more prominent, readers may judge preprints that never go on to be published in journals as flawed.
- There are no best practices for clearly presenting the history of a paper in search results.
3. Better preprint recommenders
Motivation
As more and more preprints are posted, readers need better ways to keep up to date with the current literature. Personalized recommendations could increase the visibility of preprints overall, especially those in servers that might not be considered mainstream, or in disciplines where work is distributed across many different servers. Better discovery tools would also enable preprint review and curation: feeds could help editors identify appropriate papers or reviewers, and categorization of preprints could enable automated screening tools to apply appropriate criteria based on field or type of study (for example, to only flag papers as missing an ethics statement if that is indeed a problem).
Example user stories
- As a reader, I want to see a list of relevant preprints by clicking on keywords that appear next to other preprints/articles I’m viewing so that I can find preprints relevant to my research. –Bioinformatician, Research Institution, Kenya
- As a reader (educator), I want to get an updated, curated list of preprints relevant to my coursework so that I can incorporate new science into my lectures. –Cell biologist, Primarily Undergraduate Institution, USA
- As a reader/citer, I want to get convenient (ToC-like) alerts about preprints so that I can discover preprints relevant to me. –Stem cell biologist, Research Institution, China
Desired features
- Enable introductory/onboarding feeds to combat the “empty feed” problem
- Categorize or recommend against many different axes, e.g. field, technique, or type of study
- Automatically extract terms and keywords from preprint
- Generate “More like this” recommendations
- Ensure reliable categorization of preprints to enable trust and avoid frustration
- Provide multiple ways for users to find content to add the element of serendipity
- Surface latest updates (such as withdrawals/removals, journal article retractions, and new versions) when recommending preprints
- Highlight preprints based on the presence or content of reviews
Challenges and concerns
- It is difficult or impossible to create agreement on a universal taxonomy since keywords are not understood the same way by everyone.
- Categorizing preprints does not guarantee the ability to identify preprints that will be interesting to a reader.
- Selecting the right granularity for categorization is challenging.
- Biases may be introduced from training sets and designers, which may create a skewed view of the research literature.
- Personalization creates privacy issues.
4. Summaries and badges for review content
Motivation
As the volume of feedback on preprints grows, it will become increasingly important to manage information overload. Certain types of information from assessments could be consolidated into a simplified badge or summary. In addition to preprint servers, similar information could also be displayed in search platforms and within a journal’s editorial dashboards.
User stories
- As a reader, I want to see badges or summaries of comments or feedback on preprints, so that I can understand community perceptions of a preprint’s credibility and value. –Climate policy consultant, Bangladesh
- As an author, I want to get a highly-visible badge when my preprint is reviewed (e.g., by PREreview), so that I can be rewarded for getting feedback on my preprint. –Plant scientist & microbiologist, Research Institution, South Africa
- As a reader, I want to see clear indicators of when a comment on a preprint was written by a verified individual so that I can trust comments. – Public health researcher, Research Institution, Bangladesh
Desired features
- Develop consolidated ways of displaying summary information about review on preprint servers and search engines
- Develop standardized quality metrics for preprints that can be used by reviewers
- Design badges and other review features to mitigate inequities in attention to papers based on author identity, institution, country, or other features
Challenges and concerns
- Badges may oversimplify review, be used as a crutch by readers to avoid reading the review themselves, and may become an end to themselves, recapitulating perverse incentives
- Lacking standardized definitions, the meaning of badges may be unclear to readers
- Too many badges or other indicators of review would create clutter and confusion, and would devalue their usefulness
5. Improved support for preprint review
Motivation
Preprint review can allow researchers to collaborate in new and radically different ways that were impossible to imagine in the traditional publishing system. It can take the form of structured or unstructured commenting, curation of preprints, or automated checks that would be laborious for a human to perform. Review on preprints benefits readers by adding additional context for interpreting papers, and authors can use it to improve their papers.
Many of the shared technologies discussed at the workshop were related to preprint review, and much of the discussion in these sessions focused on broad underlying issues affecting preprint review as a whole. While some of these issues could be partially addressed with technical interventions, most require social and cultural change.
Desired features
- Ensure review metadata (including persistent identifier(s)) is included (see Standardized review metadata section)
- Facilitate mining of the content of the peer reviews
- Ensure screening mechanisms don’t disproportionately flag work from under-resourced authors
- Track reviewer views to indicate impact
- Make a clear distinction between formal peer review from informal commenting
- Develop metrics of peer review quality, objectivity and depth
- Enable live conversations among reviewers about a paper
- Preserve options for reviewer pseudonymity and anonymity
- Ensure that reviews are highly visible and directly linked to preprints via metadata and PIDs
- Create systems for authors and reviewers to disclose conflicts of interest
- Develop a code of conduct for review services
- Surface journal review on preprints
- Create a central or distributed clearinghouse for preprint reviews (see Standardized review metadata below)
Challenges and concerns
- Preprint review could disrupt the “freshness” of preprints, which currently exist without labels or judgements, for both authors and readers.
- Authors may be deterred from preprinting by fear that their work will receive negative comments.
- Preprint review could displace conventional publishing, at first duplicating the effort of journals and then potentially harming them.
- Preprint servers could be harmed by the perception that they are encroaching on the role of journals, or by the confused perceptions of the role of a preprint server and how it is separate from preprint review projects.
- First-mover advantages might cause a monopoly or disadvantage under-resourced or less visible preprint servers and review entities.
- Visible reviews may dissuade participation by raising the stakes for commenting.
- It is unclear what constitutes a “review” and what should be displayed to readers as such.
- Incorrect or misleading review could create confusion or misinformation. For example, readers may be harmed if a service endorses an article later found to have serious flaws. At the same time, authors may be harmed by incorrect or unfair flagging of problems.
- Centralized solutions, as opposed to decentralized interoperable structures, tend to concentrate control over technology and/or content. This can lead to a monopoly which is not ideal for open science.
- In the absence of transparent reviewer identities or editorial moderation, the system may be “gamed” with fake reviews.
6. Preprint server integration with review platforms
Motivation
Currently, preprint review may be difficult to find since links from preprints to related reviews are not ubiquitous. In addition to visibility, other forms of integration, such as exchanging requests to review or authorizing review services to post material on behalf of authors, would streamline author experiences.
Example user stories
- As an author, I want to directly invite colleagues to read and review my work, so that I can get high-quality feedback. –Neuroscientist and meta researcher, Research Institution, Brazil
- As an author and a reviewer, I want to be able to chat directly with the reviewer/author, so that I can quickly resolve issues/questions regarding the manuscript and finish the review process quicker. –Physicist, Research Institution, Bangladesh
- As an author, I want to get a list of similar authors when I upload my preprint so that I can make connections in my field. –Agricultural economics researcher, University, Nigeria
Desired features
- Present authors with the option to request reviews (including automated review) upon preprint submission
- Display reviews next to the preprint
- Allow third parties to post a revised version of a preprint on behalf of authors
Challenges and concerns
- Preprint servers may wish to avoid encroaching on the role of journals.
- Endorsing or partnering with particular services may present challenging decisions.
7. Standardized review metadata
Motivation
Many requested features, such as notifications about comments and new versions, badges or summaries of preprint peer review, and review service integration with preprint servers, are dependent on the availability of metadata about events related to preprints. Currently, this information is heterogenous in its content and expression and is difficult to aggregate.
Aggregated, standardized metadata presented, for example, as an event stream, would enhance the visibility of preprint reviews and other services that extend the utility of preprints. It would enable readers to see reactions to preprints from diverse communities wherever the paper appears, driving trust in preprints. This would enable reviews and other information to be more easily ingested into other systems where they could be used, such as journal editorial workflows.
Desired features
- Structured descriptions of reviewing services is needed as a minimal starting point.
- The following signals should be included, along with basic metadata about the preprint in question (such as persistent identifier, title, and authors):
- New version posted
- Link to new version of the preprint
- Information about a new version of record match
- Author requests for review
- Expertise of reviewer
- Manuscript section needed
- Time limit
- Type of feedback
- Review processes
- Metadata for the review, such as:
- Persistent identifier of the review
- Date
- Author of review
- Authenticated ORCiDs for author of review
- Reference to object being reviewed (and its specific version)
- Reference to the author response if available
- Reviews obtained in the same ’round’ should be grouped together (complementary expertise, independent assessment, post-review cross-commenting)
- Licensing information on reviews
- Policies of review service
- Whether service is linked to none/one/many/any journal(s)
- Whether there is an editorial decision or recommendation after review
- Whether it is author-driven
- Whether there is a pre-selection/triage before review
- Whether service applies formal/defined guidelines for reviewers
- Preservation strategy of the review service
- Expertise/conflict of interest of reviewers
- Potentially include structured review content, enabling it to be better summarized
- Endorsement (eg from Peer Community In), decision, or scalar ranking (eg Rapid Reviews: COVID-19 uses a 1-5 scale)
- Updates on journal processes
- Metadata for the review, such as:
- Author response to review
- Reference to the review that the response refers to
- Meta review
- Curation of review feeds
- Highlighting useful review
- Potentially, author endorsements of what to display
- New version posted
Challenges and concerns
- Expecting all services to conform to a standard could result in the loss of unique features that serve individual services, resulting in an inflexible system and stagnation of innovation.
- Standards also constitute barriers for participation by raising the bar for what is minimally required.
- Focusing on a temporal order of events may shift the attention of developers away from other important aspects of the process that are not time-dependent, such grouping reviews by round or service, linking groups of reviews to a response, and capturing reviewer cross-commenting and discussion.
- A service to deliver aggregated, standardized metadata could take many forms, for example:
- Decentralized feeds from which metadata could be harvested asynchronously. A distributed system could potentially be more robust, but it could cause delays in propagation of reviews, leading to confusion for readers. (Notifications may be preferable in some areas where volume is low, such as small subject areas or tracking a small number of preprints of interest.)
- Central clearinghouse for preprint reviews. This would avoid duplication of effort (e.g., in building and maintaining APIs) but it is potentially a single point of failure and a single point of control. It would also enable screening of content prior to inclusion in the stream, which may entail governance and scaling challenges.
8. Author options for receiving reviews
Motivation
Potential reviewers (especially early career researchers or those from underrepresented groups) may be deterred by concerns that authors are not receptive to feedback on their preprint. Knowing that authors have the time and inclination to respond to and incorporate their comments would make commenting on preprints more attractive. At the same time, some authors would like to receive additional feedback, especially from reviewers with specific expertise. Requesting feedback within a certain time window (e.g., prior to journal submission or revision) would encourage public reviews to be provided when they are most useful to authors and journal editors as well.
At the same time, offering authors more control over how reviews are displayed (for example, along with their responses) could encourage adoption of a public reviewing culture.
Example user stories
- As a reviewer, I want to see an indicator when preprint authors want comments so that I can feel confident that my comments are welcomed. –Neuroscience and metascience researcher, Research Institution in Brazil
- As an (early career) reader, I want to be invited to ask questions to authors or leave comments on preprints, so that I can feel more comfortable engaging in public dialog. –Clinician / Cancer biology researcher, Research Institution, UK
- As a reviewer/teacher, I want to have a convenient way to set up conversations with authors of preprints, so that I can set up a visit to my class or provide feedback to them. –Cell biologist, Primarily Undergraduate Institution, USA
Desired features
- Allow authors to indicate:
- The section or aspect of the work they want feedback on
- The review service, society, or type of expert needed they would like feedback from
- When feedback would be appropriate (consider pinging authors every month to update or renew a request for review)
- Represent requests for review in metadata, enabling aggregation of affected preprints and increased visibility of these preprints
- Allow authors to respond to reviews, including automated checks. Authors’ responses should be displayed with the same level of prominence as the reviews.
- Allow authors to choose whether to show a review in some contexts.
- Enable third party reviewing services to post revisions and point-by-point responses on behalf of the authors
- Semi-private feedback may encourage requests for commenting. For example, some authors would appreciate the opportunity to respond to reviews before they are posted. However, allowing authors to request only private feedback would likely discourage public commenting.
Challenges and concerns
- Consensus on whether it is “fair game” to post reviews of a preprint is lacking. Authors may request that reviews be removed, complicating workflows.
- Unsolicited preprint review may alienate or overwhelm authors with too much feedback. Even if feedback is irrelevant, authors may feel compelled to spend time responding. If authors view the potential for feedback as a negative, this may discourage them from posting preprints in the first place.
- Reviewers may feel that their time is wasted if their comments are not incorporated by authors.
- The absence of a request for feedback may be seen as the author having no interest for feedback, when in fact authors could just be unaware of the system.
- Low adoption of such a system may also reinforce the idea that most authors don’t want comments, even if they do.
- Because negative reviews are typically hidden from readers, negative reviews on preprints may unfairly punish authors, making their work seem proportionally more poorly-received than journal articles.
- Requests for review are time-dependent. If authors request feedback but don’t indicate when they are closed to feedback (e.g., when a paper is published), this may waste the time of reviewers.
- There may be inequities or biases in what review services choose to review.
- This functionality could be integrated with several different services. Integrating with preprint servers would allow authors to make requests when they submit, but preprint servers may wish to avoid being perceived as playing the role of a journal. Integration with preprint reviewing services could trigger a template to be prefilled or contact users of the service with appropriate expertise.
9. Notifications and updates
Motivation
Most preprints are updated in some way, either through the authors posting a new version to the server or publishing the paper in a journal, posing a challenge for readers. Many preprints also receive comments and reviews that authors may be unaware of. A notification service would enable willing authors and other interested parties (for example, those who have cited or saved a preprint to their library) to receive an alert when preprints are either updated or commented on. This would help readers stay on top of changing literature and enable authors to more rapidly receive and respond to comments on preprints.
Example user stories
- As an author, I want to get email alerts or notifications when someone leaves a comment on my article so that I can get constructive criticism to help improve my article or identify new collaborators. –Neuroscience and molecular biology researcher, Research Institution, Spain
- As a reader, I want to get alerts when preprints are updated or published, containing a summary of how they’ve changed so that I can keep up with the literature. –Cell biologist, Research Institution, India
- As a reader, I want to get a notification that a preprint I’ve cited or saved to my library is now published so that I can stay up to date with the literature. –Cell biologist & neuroscientist, Research Institution, Chile
Desired features
- Allow subscribers to customize notification subscriptions and frequency
- Get notifications when:
- New version of preprint is posted
- Journal article match occurs
- New comment, review, news mention, or citation is received
Challenges and concerns
- Frequent, unsolicited notifications may be perceived as spam
10. Other needs
Commenting and endorsements as highlights, annotations, proposed changes
Motivation
Enabling readers to comment on preprints in-line through annotations can lower the barrier to participating in peer review. “Crowd” review from many different scientists leaving small comments on the parts of papers closest to their own expertise would be more robust than traditional peer review, and may lead to consensus since it is conducted in the open. Since annotations are more firmly anchored to a specific part of a manuscript, they have the opportunity to be more actionable and constructive than traditional peer review, helping authors to more rapidly revise their paper. Annotations would help readers combat information overload by drawing attention to the most interesting and controversial parts of an article, and they could also be used to annotate individual sentences with links to data repositories.
User stories
- As an author, I want to see feedback on my article as in-line annotations so that I can more easily improve my paper. –Social scientist, Research Institution, Mexico
- As an author, I want to allow readers to propose changes, recalculate figures, and interact with code in manuscripts I post as Jupyter notebooks, R markdown, Shiny apps, etc., so that I can collaborate with readers. –Bioinformatician, Research Institution, Kenya
- As an author, I want to have readers propose comments and changes to my paper in-line (similar to GitHub), so that I can improve my paper and find new collaborators. –Bioinformatician, Research Institution, Estonia
Integration with authoring tools
Motivation
Integration with authoring tools would not only allow authors to generate native XML or HTML preprints, reaping the benefits described in the markup language conversion section, but would also give them more direct control over formatting and presentation and encourage the integration of more interactive features.
User stories
- As an author, I want to submit manuscripts created by an HTML/XML authoring tool directly to my chosen preprint server, so that I can create a rich preprint with hyperlinks and embedded videos and also data. –Agricultural researcher, Research Institution, India
- As an author, I want to easily create standardized manuscripts, so that I can post professional-looking preprints. –Physicist, Research Institution, Bangladesh
Clearly labeling preprints as preprints
Motivation
As the number of preprint servers grows and they are increasingly cited along with journal articles in news media, the scholarly literature, and elsewhere, readers wish to know the peer review status of the article in question. This could be achieved with clear labeling on preprint servers, by journalists, in search engines, and in citation styles.
User stories
- As a (non-specialist) reader, I want to see clear disclaimers about what a preprint is, so that I can better understand the status of preprints. –Synthetic biologist, Community biology setting, USA
- As a reader, I want to distinguish very clearly that the manuscript I am reading is actually a preprint, so that I can use it accordingly. –Social scientist, Research Institution, Mexico
- As a reader, I want to see preprints clearly (automatically) labeled in reference lists, so that I can approach the article with appropriate skepticism. –Public health researcher, Research Institution, Bangladesh
Lay summaries
Motivation
Readers desire accessible summaries of papers. These summaries could also help more general readers avoid drawing mistaken conclusions from preprints by enabling the authors to “prebunk” misinterpretations.
User stories
- As a (clinician) reader, I want to see digests and summaries of preprints, so that I can keep up with the literature. –Clinician / Cancer biologist, Research Institution, UK
- As a reader, I want to read an accessible summary of preprints, so that I can become aware of interesting preprints. –Stem cell biologist, Research Institution, China
- As a (non-specialist) reader, I want to read a lay summary and see a graphic abstract accompanying preprints, so that I can better understand the preprint. –Synthetic biologist, Research Institution / Community biology setting, USA
Advanced search features for preprints
Motivation
Both awareness and coverage of existing search tools and alert features could be improved. At the same time, more fundamental deficits in metadata (such as unambiguous identification of authors or institutions) could be addressed by making the submission process more stringent.
User stories
- As a reader, I want to use the same advanced search features (e.g., boolean operators) for preprints as are available to me for journals, so that I can include preprints in meta-analysis. –Neuroscientist and meta researcher, Research Institution, Brazil
- As a reader, I want to search and/or get alerts about preprints from particular authors, labs, or institutes, so that I can find preprints relevant to my research. –Evolutionary biologist, Research Institution, India
- As a reader, I want to use advanced search and email/alert features (as in PubMed) to find preprints, so that I can find preprints relevant to me. –Synthetic biologist, Community biology setting, USA
Cross-platform search or integration
Motivation
While usage of preprint servers varies among disciplines, content may be spread across community and publisher servers. Indexing of all relevant preprint servers in search tools and databases is incomplete. Furthermore, authors may wish to ensure their preprint reaches the maximum possible audience, and so desire their preprints to appear in the on-site search tools at more than one server.
User stories
- As a reader, I want to search for all preprints in one place, so that I can find research regardless of what server it is posted on. –Evolutionary genetics researcher, Research Institution, India
- As an author, I want to duplicate my manuscript across several repositories (with appropriate metadata) so that I can ensure that it is not lost. –Agricultural researcher, Research Institution, India
- As a reader, I want to have an easy way to search across multiple repositories, so that I can be aware of all relevant preprints. –Cell biologist, Primarily Undergraduate Institution, USA
Embedding video, code and other media types
Motivation
As digital objects, preprints have the potential to integrate content that can’t fit into a traditional paper journal article. Preprints could re-envision the article’s relationship with data, code, and video, integrating these elements inline rather than referencing external supplements.
User stories
- As an author, I want to allow readers to propose changes, recalculate figures, and interact with code in manuscripts I post as Jupyter notebooks, R markdown, Shiny apps, etc, so that I can collaborate with readers. –Bioinformatics researcher, Research Institution, Kenya
- As a (grassroots scientist) author, I want to share my science through video and other non-traditional formats, so that I can communicate and share quickly. –Synthetic biologist, Research Institution / Community biology setting, USA
- As an author, I want to be able to embed SBOL files in my preprint, so that others can reuse my synthetic biology parts. –Synthetic biologist, Research Institution / Community biology setting, USA
Meta review
Motivation
As the volume of reviews of preprints grows, filtering and highlighting useful review will become increasingly important. This selection may be performed by selecting individual review services for indexing or display or by moderating or rating individual reviews.
Desired features
- Curation of review feeds
- Highlighting useful review
- Enable service providers to make decisions about how to screen or curate sources of preprint review. Possible approaches:
- Display only review content requested or endorsed by authors.
- Display only reviews from trusted services or platforms, which set expectations for reviewers and perform their own moderation.
- Display reviews based on the credentials of individual reviewers and the content of their reviews.
Challenges and concerns
- There is a gray area between insubstantial comments and detailed reviews. It is unclear what constitutes a “review” and what should be displayed to readers as such.
- Moderation of reviews is time consuming, but necessary to remove ad hominem attacks or other unprofessional content, which could dissuade readers from paying attention to preprint review more generally.
- Anonymous commenting might be seen as less trustworthy than traditional peer review.
- Service providers will need to make decisions about how to screen or curate sources of preprint review. There may be several approaches to this problem:
- Display only review content requested by authors.
- Display only reviews from trusted services or platforms, which set expectations for reviewers and perform their own moderation.
- Display reviews based on the credentials of individual reviewers and the content of their reviews.
- Decisions on what sources of review to consider “trustworthy” may serve select stakeholders or communities while neglecting the needs of others.
Acknowledgements
While the authors are solely responsible for this summary, the information within originated from many stakeholders. We thank many anonymous contributors as well as: Rich Abdill, Anurag Acharya, Michele Avissar-Whiting, Anita Bandrowski, Nokome Bentley, Rachel Besen, Matt Bettonville, Damian Bird, Denis Bourguet, Rachel Burley, Jennifer Byrne, Guillaume Cabanac, Clarissa Franca Dias Carneiro, Andy Collings, Humberto Debat, Tom Demeranville, Vilas Dhar, Jim Entwood, Gunther Eysenbach, Ashley Farley, Katie Funk, Joe Gorney, Kate Grabowski, Thomas Guillemaud, Emily Gurley, Sridhar Gutam, Gabriel Harp, M Tasdik Hasan, Chelsea Haupt, Jo Havemann, Daniel Himmelstein, Samantha Hindle, Michele Ide-Smith, John Inglis, Takeshi Irie, Amit Jain, Alexander Ketchakmadze, Caleb Kibet, Veronique Kiermer, Robert Kiley, Martin Klein, Sebastian Kohlmeier, Cyril Labbé, Michael Lacy, Rachael Lammey, Thomas Lemberger, Jennifer Lin, Nick Lindsay, Ivonne Lujano, Giuliano Maciocci, Gabriele Marinello, Jo McEntyre, Heather McKay, Alex Mendonça, Priscilla Mensah, Mayukh Mondal, Thabiso Motaung, Ross Mounce, Kleber Neves, Josh Nicholson, Michèle Nuijten, Louise Page, Mate Palfy, Michael Parkin, Damian Pattinson, Alex Pearlman, Nici Pfeiffer, Eleonora Presani, Iratxe Puebla, Omar Quintero-Carmona, Kristen Ratan, Nic Riedel, Martyn Rittman, Andy Robinson, John Sack, Daniela Saderi, Richard Sever, Paul Shannon, Kathleen Shearer, Adiran Stanley, Bodo Stern, Katherine Szarama, Kaitlin Thaney, Raphaël Tournoy, Lilian Treasure, EJ van Lanen, Tim Vines, Christina Vizcarra, Alex Wade, Susan Walsh, Tracey Weissgerber, and Dan Whaley.
Funding for this project was provided by the Chan Zuckerberg Initiative. We thank Patricia Flores and Grady Fike for helping produce and publish this report.
How to cite this work
Jessica Polka, Carly Strasser, and Dario Taraborelli (2021). Shared technology needs for preprints (Version 1.1). Zenodo. https://doi.org/10.5281/zenodo.4700569
“Shared technology needs for preprints” by Jessica Polka, Carly Strasser, and Dario Taraborelli is licensed under CC BY 4.0
Appendix
Project overview and methods
The project began with an information-gathering phase in which we collected two parallel streams of information: interviews with users of preprints and consultations with leads of preprint-related projects. We grouped the needs emerging from these processes together to form a list of prioritized technical needs which were then selected, by interest level, by participants in a workshop held on February 2, 2021 as the basis for discussion in breakout sessions. We distilled themes from those discussions into the major sections outlined in this report. Each step is described in greater detail in the sections to follow.
Figure 1. Overview of project phases
User interviews
Goals
We sought to identify unmet technological needs that could promote preprint adoption among varied communities of preprint users, beyond those we typically encounter through our work in a primarily North American, computationally-focused context.
Methodology
We conducted 30-minute video or phone interviews with [30] participants, representing three categories:
- Countries underrepresented in preprint use relative to overall scientific output (see Abdill et al.): India, Bangladesh, Spain, Estonia, Kenya, Nigeria, Mexico, Brazil, Chile
- Academics at Primarily Undergraduate Institutions (PUI) and Minority Serving Institutions (MSI) in the US
- Those outside of a basic science, academic research context: clinicians, community bio, bioethics, pharma
Before each interview, we collected information about the participant’s research and institutional context. During the interview, we provided some information about the project and process, especially its focus on technology rather than social or policy change. We then asked the following questions, time permitting, with occasional follow ups:
- What is your level of experience with preprints? Have you read, posted, cited, commented?
- Would you say this level of experience is typical for people in your institution/field?
- Are there any reasons (besides low visibility, lack of recognition, and unsupportive policies) that preprints aren’t more widely used?
- Describe what your ideal preprint experience would be, from the perspective of posting, reading, citing, or reviewing.
- Are there any missing tools or features that would make preprints more appealing to post, read, cite, or review?
- Are there any situations in which you’d look preferentially at preprints instead of journal articles?
We took notes during the interview, which we cleaned up afterwards (sometimes disrupting chronology by grouping related content together). We then created user stories based on the notes. Participants had a chance to review, edit, or redact content from the notes and user stories before sharing with CZI.
Results
The interviews yielded 96 user stories (see Table 1). We tagged these user stories with features requested. One story represented two features and is represented twice in the table below.
A recurring theme from the consultations not represented in these stories is that technology is not the main barrier to increasing preprint use. Nevertheless, many users desired improvements in preprint discovery, and less commonly, preprint authoring and submission.
Consultations with project leads
Goals
We sought to identify technological barriers encountered by preprint-related projects, preprint servers, and other infrastructure providers in the delivery of features most in demand among their user communities.
Methodology
We sent willing participants individual Google docs containing a written questionnaire. Questions varied according to the category of project being consulted. We invited all respondents to use formatting to mark responses that would like to not be shared beyond CZI/ASAPbio.
For preprint servers
- What are the enrichments, content transformations and collaborative features that your audience values the most?
- Are you able to provide support for this functionality directly through your platform?
- Are there third party services or platform integrations that provide valuable functionality for your audience based on contents and metadata from your server?
- What data are you providing to 3rd parties (such as metadata to Crossref or usage data via an API, etc)? In what format and by what means?
- Are there other data you’d like to provide to these parties, but can’t?
- What are the barriers to providing or ingesting these data?
- What features would you like to provide to users, but can’t?
- What are the barriers (technical, logistical, financial) to offering those new functionalities?
For preprint-related projects
- What does your project do for users and how does it interact with preprints?
- What is the size of your audience (active users/month)?
- What data sources and services relevant to preprints are you currently using for the project?
- How are you making your own data available to 3rd parties or other tools?
- What are some functionalities that are in high demand among your existing or potential users/contributors, but you can’t offer yet? Feel free to be creative and speculative.
- What are the barriers (technical, logistical, financial) to offering those new functionalities?
- What are the barriers (technical, logistical, financial) to the maintenance or day-to-day operation of your project?
For infrastructure providers
- How are you ingesting information provided by preprint servers? In what format and by what means?
- Are you ingesting information provided by 3rd party peer review or other overlay services? In what format and by what means?
- What data are you making available to others? In what format and by what means?
- Are there additional features regarding preprints you’d like to provide to users, but can’t? Feel free to be creative and speculative.
- Are there other data you’d like to provide to 3rd parties, but can’t?
- What are the barriers (technical, logistical, financial) to offering those new functionalities?
- What are the barriers (technical, logistical, financial) to the maintenance or day-to-day operation of your project?
We also provided each participant with a list of projects on our radar and requested suggestions for additional contacts.
Results
We received 33 completed consultation documents. We then tagged the responses to these questionnaires with a list of features derived de novo.
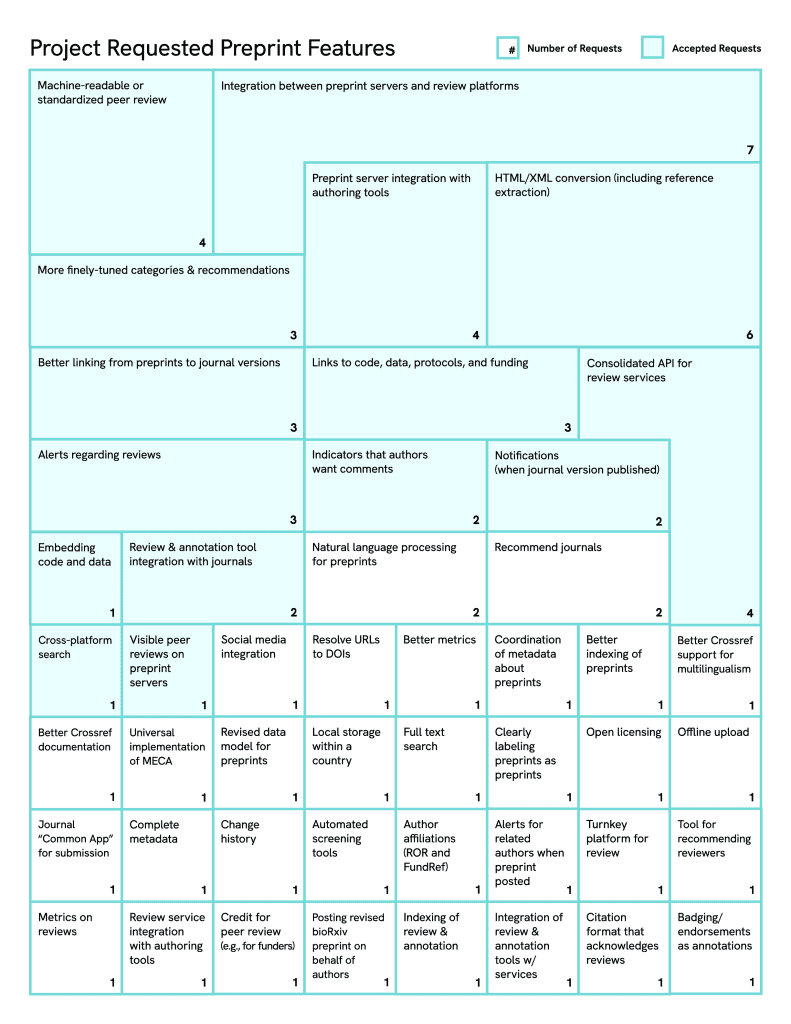
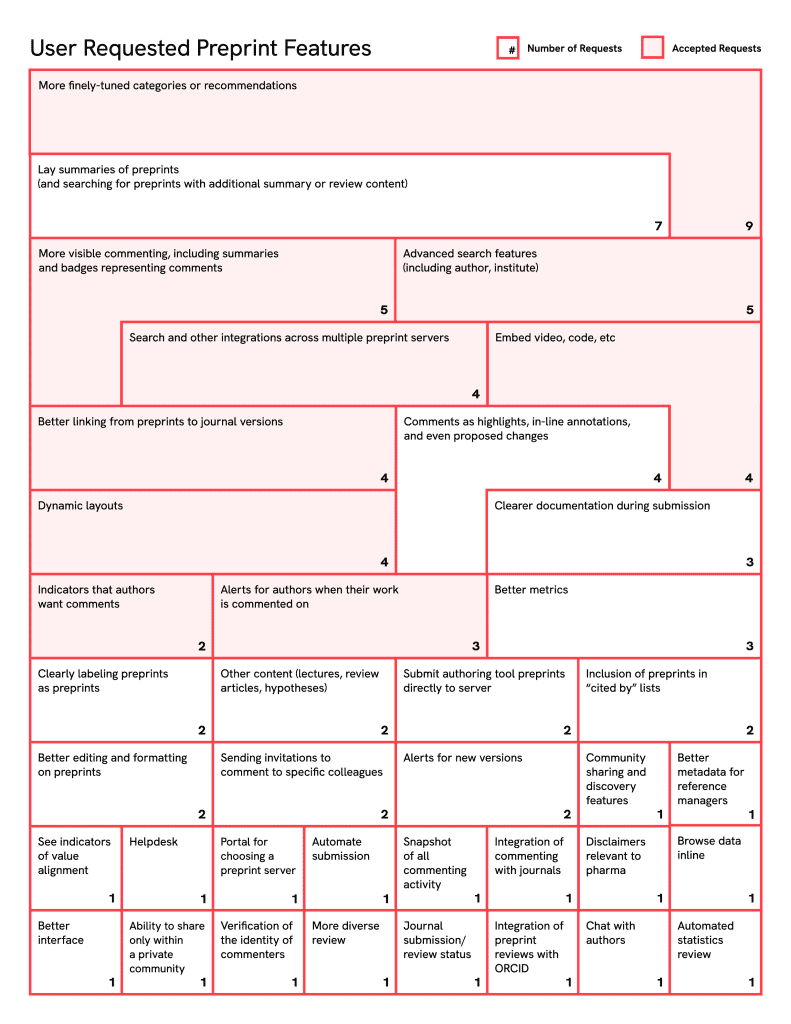
Figure 2. Consolidated features emerging from user stories and project consultations
Workshop
Goals
Equipped with a prioritized list of desired features, the goal of our workshop was to better understand, through discussion among preprint service and project providers, the shared technical needs that would best serve their audience. Through these discussions, we aimed to identify opportunities to address emerging user needs in the preprint ecosystem.
Methodology
We invited project leads and additional stakeholders (such as funders and others with deep knowledge of the preprint ecosystem) to participate in a 3-hour long workshop on February 2, 2021. Upon registration, we presented workshop attendees with a list of topics ranked highly in user interviews and project consultations, enabling them to indicate their level of interest in each. We selected the top 9 of these, duplicating the most highly-ranked session (Integration between preprint servers and review platforms) so that more attendees could participate.
For each session, we invited one attendee who indicated strong interest in the topic to act as facilitator. Prior to the meeting, we provided each facilitator with a briefing document containing pertinent user stories and excerpts from project consultations. During the breakout sessions, facilitators used this information to provide attendees with a brief introduction to the problem before introducing a silent writing exercise and moderating an ensuing discussion, according to the prompts provided below.
Breakout session agenda
5’ – Facilitator gives an overview of the proposed need with related user stories and examples from ASAPbio/CZI consultation
10’ – Silent writing
- If this technology existed, how would it benefit the audience or community of your project/organization?
- What would your project/organization do with it?
- Are there unintended consequences of adopting this technology?
45’ – Discussion
- Read through one another’s statements – do any themes emerge? What’s missing?
- What would be needed to reduce barriers for projects to take advantage of this technology?
- What existing technology/organization comes close to solving the problem? Could it be expanded or supported? Are there organizations working on the problem not represented here who should be brought into the conversation?
Table 1. User stories
| ID | Feature | User type | Story feature | Goal | Country | Field | Context |
|---|---|---|---|---|---|---|---|
| 1 | Dynamic layouts | As a reader, I want to | view articles in different layouts (figures on the side, etc), so that I can | more easily cross-reference figures and results | India | Evolutionary genetics | Research Institution |
| 2 | Searching/integrations across multiple preprint servers | As a reader, I want to | search for all preprints in one place, so that I can | find research research regardless of what server it is posted on | India | Evolutionary genetics | Research Institution |
| 3 | Better linking to journal versions | As a reader, I want to | reliably see links on preprints to their journal version, so that I can | keep up to date with research | India | Evolutionary genetics | Research Institution |
| 4 | Alerts for authors when their work is commented on | As an author, I want to | get email alerts or notifications when someone leaves a comment on my article, so that I can | get constructive criticism to help improve my article or identify new collaborators | Spain | Neuroscience and molecular biology | Research Institution |
| 5 | More finely-tuned categories or recommendations | As a reader, I want to | get alerts about preprints based on keywords or AI curation, so that I can | stay up to date on research relevant to me | Spain | Neuroscience and molecular biology | Research Institution |
| 6 | Clearer documentation during submission | As an author, I want to | get clear instructions about journal requirements (eg manuscript format) when transferring my paper from a preprint server to a journal, so that I can | transfer my paper more easily. | Brazil | Neuroscience, metascience | Research Institution |
| 7 | More finely-tuned categories or recommendations | As a reader, I want to | use improved search tools that behave as expected in response to keywords or more finely-tuned categories, so that I can | find preprints relevant to me. | Brazil | Neuroscience, metascience | Research Institution |
| 8 | Advanced search features (including author, institute) | As a reader, I want to | use the same advanced search features (eg boolean operators) for preprints as are available to me for journals, so that I can | include preprints in meta-analysis. | Brazil | Neuroscience, metascience | Research Institution |
| 9 | Integration of preprint reviews with ORCID | As a reviewer, I want to | have my reviews of preprints appear in my ORCID profile, so that I can | get credit for preprint reviewing activity. | Brazil | Neuroscience, metascience | Research Institution |
| 10 | An indicator that authors want comments | As a reviewer, I want to | see an indicator when preprint authors want comments, so that I can | feel confident that my comments are welcomed. | Brazil | Neuroscience, metascience | Research Institution |
| 11 | More finely-tuned categories or recommendations | As a reader, I want to | access a curated list of preprints related to bioinformatics software, so that I can | find preprints relevant to my research. | Kenya | Bioinformatics | Research Institution |
| 12 | Inclusion of preprints in “cited by” lists | As a reader, I want to | see a list of preprints that cite an article I’m looking at, so that I can | find preprints relevant to my research. | Kenya | Bioinformatics | Research Institution |
| 13 | More finely-tuned categories or recommendations | As a reader, I want to | see a list of relevant preprints by clicking on keywords that appear next to other preprints/articles I’m looking at, so that I can | find preprints relevant to my research. | Kenya | Bioinformatics | Research Institution |
| 14 | Better editing and formatting on preprints | As a reader, I want to | read preprints that are edited and formatted, so that I can | have a smoother reading experience. | Kenya | Bioinformatics | Research Institution |
| 15 | Submit HTML/XML authoring tool preprints directly to a chosen server | As an author, I want to | submit manuscripts created by an HTML/XML authoring tool directly to my chosen preprint server | so that I can create a rich preprint with hyperlinks and embedded videos and also data | India | Agriculture | Research Institution |
| 16 | Better metadata for reference managers | As a reader, I want to | have better preprint metadata harvested by reference managers | so that I can appropriately include preprints in my reference library | India | Agriculture | Research Institution |
| 17 | Searching/integrations across multiple preprint servers | As an author, I want to | duplicate my manuscript across several repositories (with appropriate metadata) | so that I can ensure that it is not lost | India | Agriculture | Research Institution |
| 18 | Inclusion of preprints in “cited by” lists | As a preprint server lead, I want to | know where preprints in my server are being cited | so I can understand the impact of preprints | India | Agriculture | Research Institution |
| 19 | More visible commenting, including summaries and badges representing comments | As a reader, I want to | see badges or summaries of comments or feedback on preprints, so that I can | understand community perceptions of a preprint’s credibility and value | Bangladesh | Climate | Policy |
| 20 | Advanced search features (including author, institute) | As a reader, I want to | search and/or get alerts about preprints from particular authors, labs, or institutes, so that I can | find preprints relevant to my research | India | Evolutionary Biology | Research Institution |
| 21 | Lay summaries of preprints (and searching for preprints with additional summary or review content) | As a reader, I want to | read a justification for why authors selected the category/subject area, so that I can | find preprints relevant to my research | India | Evolutionary Biology | Research Institution |
| 22 | Dynamic layouts | As a reader, I want to | see figures interspersed in the text of preprints, so that I can | have a smoother reading experience | India | Evolutionary Biology | Research Institution |
| 23 | Disclaimers relevant to pharma | As an author or supporting medical writer, I want to | share my preprint with a disclaimer that it is intended as scientific exchange, so that I can | comply with laws surrounding off-label promotion of drugs | UK | Biomedical | Pharma/Medical writing |
| 24 | Comments as highlights, in-line annotations, and even proposed changes | As an author, I want to | See feedback on my article as in-line annotations, so that I can | more easily improve my paper | Mexico | Social sciences | Research Institution |
| 25 | Snapshot of all commenting activity | As an author, I want to | Show someone a snapshot of all the reviewing and commenting activity on my preprint, so that I can | get credit and recognition from evaluators (PhD program faculty) | Mexico | Social sciences | Research Institution |
| 26 | Clearly labeling preprints as preprints | As a reader, I want to | distinguish very clearly that the manuscript I am reading is actually a preprint, so that I can | use it accordingly | Mexico | Social sciences | Research Institution |
| 27 | Alerts for authors when their work is commented on | As an author, I want to | get notifications when my article is commented on, so that I can | benefit from feedback on my preprint | South Africa | Plant science, microbiology | Research Institution |
| 28 | Help choosing a preprint server through a centralized portal | As an author, I want to | submit my preprint to a centralized system, so that I can | select the best server | South Africa | Plant science, microbiology | Research Institution |
| 29 | More visible commenting, including summaries and badges representing comments | As an author, I want to | get a highly-visible badge when my preprint is reviewed (eg by PREreview), so that I can | be rewarded for getting feedback on my preprint | South Africa | Plant science, microbiology | Research Institution |
| 30 | Clearer documentation during submission | As an author, I want to | see “model preprints” that have been cited or undergone peer review, so that I can | understand the potential of preprints | South Africa | Plant science, microbiology | Research Institution |
| 31 | Lay summaries of preprints (and searching for preprints with additional summary or review content) | As a reader, I want to | Discover research and relevant bioethics work together, so that I can | see research in its ethical context | USA | Bioethics | Community bio |
| 32 | Lay summaries of preprints (and searching for preprints with additional summary or review content) | As a (nonspecialist) reader, I want to | Read an author-provided lay summary of a preprint, so that I can | better understand the content I’m interested, but not fluent in | USA | Bioethics | Community bio |
| 33 | Lay summaries of preprints (and searching for preprints with additional summary or review content) | As a reader, I want to | read author statements on potential dangers or implications to society, so that I can | be assured authors have considered ethical implications of their work | USA | Bioethics | Community bio |
| 34 | Better interface | As a (nonspecialist) reader, I want to | browse preprint servers with a more user-friendly interface, so that I can | access preprints more easily | USA | Bioethics | Community bio |
| 35 | More finely-tuned categories or recommendations | As a reader, I want to | browse preprints by my sub-discipline, so that I can | find preprints relevant to me | Nigeria | Agricultural economics | University |
| 36 | Lay summaries of preprints (and searching for preprints with additional summary or review content) | A a reader, I want to | read short jargon-free summaries of preprints, so that I can | find preprints relevant to me | Nigeria | Agricultural economics | University |
| 37 | A simple way for authors to send invitations to comment to specific colleagues | As an author, I want to | get a list of similar authors when I upload my preprint, so that I can | make connections in my field | Nigeria | Agricultural economics | University |
| 38 | Ability to share only within a private community | As an author, I want to | share my preprint only with people who register to be a part of a community, so that I can | have a sense of ownership and security | Nigeria | Agricultural economics | University |
| 39 | Comments as highlights, in-line annotations, and even proposed changes | As an author, I want to | see reader highlights and in-line comments, so that I can | improve my paper | Kenya | Bioinformatics | Research Institution |
| 40 | More finely-tuned categories or recommendations | As a reader, I want to | get recommendations from AI or other forms of categorization, so that I can | find preprints relevant to me | Kenya | Bioinformatics | Research Institution |
| 41 | Metrics | As an author | see more information about readers of my preprint over time, so that I can | understand the impact of my work | Kenya | Bioinformatics | Research Institution |
| 42 | Comments as highlights, in-line annotations, and even proposed changes | As an author, I want to | allow readers to propose changes, recalculate figures, and interact with code in manuscripts I post as Jupyter notebooks, R markdown, Shiny apps, etc, so that I can | collaborate with readers | Kenya | Bioinformatics | Research Institution |
| 42 | Embed video, code, etc | As an author, I want to | allow readers to propose changes, recalculate figures, and interact with code in manuscripts I post as Jupyter notebooks, R markdown, Shiny apps, etc, so that I can | collaborate with readers | Kenya | Bioinformatics | Research Institution |
| 43 | Other types of narrative content (lectures, review articles, negative results, hypotheses) | As an author, I want to | quickly and easily publish my negative or less interesting results, so that I can | make my work citable and help others | Kenya | Bioinformatics | Research Institution |
| 44 | Automated statistics review | As a reader, I want to | see preprints that have undergone an automated statistics check, so that I can | know that what I am reading is sound | Japan | Neuroscience | Research Institution |
| 45 | Other types of narrative content (lectures, review articles, negative results, hypotheses) | As an author, I want to | post hypotheses, so that I can | share my ideas with the community | Japan | Neuroscience | Research Institution |
| 46 | Advanced search features (including author, institute) | As a reader, I want to | use PubMed and Google Scholar advanced search and alert features to find preprints, so that I can | find preprints relevant to me | India | Cell biology | Research Institution |
| 47 | Alerts for new versions | As a reader, I want to | get alerts when preprints are updated or published, containing a summary of how they’ve changed, so that I can | keep up with the literature | India | Cell biology | Research Institution |
| 48 | More diverse review | As a reviewer, I want to | be involved in projects that provide constructive feedback on preprints, so that I can | avoid recreating the geographical biases of traditional peer review | India | Cell biology | Research Institution |
| 49 | Alerts for authors when their work is commented on | As an author, I want to | Get notifications when someone comments on my preprint, so that I can | Get feedback on my work | Chile | Cell biology/neuroscience | Research Institution |
| 50 | More finely-tuned categories or recommendations | As a reader, I want to | Get emails about preprints related to my chosen keywords, so that I can | Find and comment on work that is interesting to me | Chile | Cell biology/neuroscience | Research Institution |
| 51 | Alerts for new versions | As a reader, I want to | Get a notification that a preprint I’ve cited or saved to my library is now published, so that I can | Stay up to date with the literature | Chile | Cell biology/neuroscience | Research Institution |
| 52 | A simple way for authors to send invitations to comment to specific colleagues | As an author, I want to | directly invite colleagues to read and review my work, so that I can | get high-quality feedback | Brazil | Neuroscience/metascience | Research Institution |
| 53 | A simple way for authors to send invitations to comment to specific colleagues | As an author, I want to | share my work with a group of colleagues working in my subfield, so that I can | get the right audience for my preprint | Brazil | Neuroscience/metascience | Research Institution |
| 54 | Metrics | As a reader, I want to | find the work that the researchers in my field are discussing and judging to be relevant, so that I can | get a grasp of the consensus | Brazil | Neuroscience/metascience | Research Institution |
| 55 | Better editing and formatting on preprints | As a (student) reader, I want to | download a pdf of a preprint in a compact, easy-to-read format, so that I can | have a smoother reading experience and stay within my university printing quota | USA | Cell biology | PUI |
| 56 | Lay summaries of preprints (and searching for preprints with additional summary or review content) | As a reader (educator), I want to | search for preprints with news & views or other summaries, so that I can | find material that will be helpful to to my students | USA | Cell biology | PUI |
| 57 | Searching/integrations across multiple preprint servers | As a reader, I want to | get alerts (keywords, cited by) about preprints related from multiple servers, so that I can | find relevant preprints | USA | Cell biology | PUI |
| 58 | More visible commenting, including summaries and badges representing comments | As a reader (educator), I want to | see comments from colleagues under a paper, so that I can | identify experimental design questions for my students’ journal club | USA | Cell biology | PUI |
| 59 | More finely-tuned categories or recommendations | As a reader (educator), I want to | get an updated, curated list of preprints relevant to my coursework, so that I can | incorporate new science into my classroom | USA | Cell biology | PUI |
| 60 | Lay summaries of preprints (and searching for preprints with additional summary or review content) | As a reader (educator), I want to | see a lay summary and author highlights of most important figure/figure panels, so that I can | more easily use the paper in an undergraduate journal club | USA | Cell biology | PUI |
| 61 | Clearly labeling preprints as preprints | As a reader, I want to | see preprints clearly (automatically) labeled in reference lists, so that I can | approach the article with appropriate skepticism | Bangladesh | Public health | Research Institution |
| 62 | Integration of commenting with journals | As an author, I want to | send comments I receive of my preprints to the journal reviewing my paper, so that | feedback I receive on preprints can be incorporated into the journal peer review process | Bangladesh | Public health | Research Institution |
| 63 | More visible commenting, including summaries and badges representing comments | As a reader, I want to | see feedback on preprints more prominently, so that I can | put the preprint into context | Bangladesh | Public health | Research Institution |
| 64 | Verification of the identity of commenters | As a reader, I want to | see clear indicators of when a comment on a preprint was written by a verified individual, so that I can | trust comments | Bangladesh | Public health | Research Institution |
| 65 | Metrics | As an author, I want to | have citations on my preprint and subsequent journal article pooled in all databases (including WoS), so that I can | get credit for my impact | India / Estonia | Bioinformatics | Research Institution |
| 66 | Comments as highlights, in-line annotations, and even proposed changes | As an author, I want to | have readers propose comments and changes to my paper in-line (similar to GitHub), so that I can | improve my paper and find new collaborators | India / Estonia | Bioinformatics | Research Institution |
| 67 | Lay summaries of preprints (and searching for preprints with additional summary or review content) | As a (clinician) reader, I want to | see digests and summaries of preprints, so that I can | keep up with the literature | UK | Clinician / Cancer biology | Research Institution |
| 68 | An indicator that authors want comments | As an (early career) reader, I want to | be invited to ask questions to authors or leave comments on preprints, so that I can | feel more comfortable engaging in public dialog | UK | Clinician / Cancer biology | Research Institution |
| 69 | Dynamic layouts | As a reader, I want to | see supplemental material (such as movies) inline as I’m reading a preprint, so that I can | have easier access to all relevant data. | USA | Cell biology | PUI |
| 70 | An indicator that authors want comments | As a reviewer/teacher, I want to | have a convenient way to set up conversations with authors of preprints, so that I can | set up a visit to my class or provide feedback to them. | USA | Cell biology | PUI |
| 71 | Searching/integrations across multiple preprint servers | As a reader, I want to | have an easy way to search across multiple repositories, so that I can | be aware of all relevant preprints. | USA | Cell biology | PUI |
| 72 | Submit HTML/XML authoring tool preprints directly to a chosen server | As an author, I want to | easily create standardized manuscripts, so that I can | post professional-looking preprints | Bangladesh | Physics | Research Institution |
| 73 | Advanced search features (including author, institute) | As a reader, I want to | retrieve different types of content based on search terms (lectures, review articles, research articles) so that I can | find resources that fit my needs | Bangladesh | Physics | Research Institution |
| 74 | Comments as highlights, in-line annotations, and even proposed changes | As a reader, I want to | choose to see annotations and comments of previous readers on a preprint, so that I can | better understand the paper | Bangladesh | Physics | Research Institution |
| 75 | Chat with authors | As an author and a reviewer, I want to | be able to chat directly with the reviewer / author, so that I can | quickly resolve issues / questions regarding the manuscript and finish the review process quicker | Bangladesh | Physics | Research Institution |
| 76 | Better linking to journal versions | As an author, I want to | have robust, automatic linking from my preprint to the published journal article, so that I can | help readers discover the updated version of the paper | USA | Bioengineering | MSI |
| 77 | Embed video, code, etc | As a (grassroots scientist) author, I want to | share my science through video and other non-traditional formats, so that I can | communicate and share quickly | USA | Synthetic biology, grassroots biology | Research Institution, Community bio |
| 78 | Automate submission | As a (grassroots scientist) author, I want to | upload in as few clicks as possible, so that I can | communicate and share quickly | USA | Synthetic biology, grassroots biology | Research Institution, Community bio |
| 79 | See indicators of value alignment | As a reader, I want to | know that the author or preprint server has values that I align with, so that I can | find research relevant to me | USA | Synthetic biology, grassroots biology | Research Institution, Community bio |
| 80 | Advanced search features (including author, institute) | As a reader, I want to | Use advanced search and email/alert features (as in PubMed) to find preprints, so that I can | find preprints relevant to me | USA | Synthetic biology | Community bio |
| 81 | Lay summaries of preprints (and searching for preprints with additional summary or review content) | As a (non-specialist) reader, I want to | see a lay summary of preprints, so that I can | better understand the science | USA | Synthetic biology | Community bio |
| 82 | Clearly labeling preprints as preprints | As a (non-specialist) reader, I want to | see clear disclaimers about what a preprint is, so that I can | better understand the status of preprints | USA | Synthetic biology | Community bio |
| 83 | Better linking to journal versions | As a reader, I want to | see a link to the journal version from the preprint (once published), so that I can | keep up with any updates | USA | Synthetic biology | Community bio |
| 84 | More finely-tuned categories or recommendations | As a reader/citer, I want to | get convenient (ToC-like) alerts about preprints, so that I can | discover preprints relevant to me | China | Stem cell biology | Research Institution |
| 85 | Lay summaries of preprints (and searching for preprints with additional summary or review content) | As a reader, I want to | read an accessible summary of preprints, so that I can | become aware of interesting preprints | China | Stem cell biology | Research Institution |
| 86 | Community sharing and discovery features | As a reader, I want to | discover and share preprints in an online community, so that I can | become aware of interesting preprints | China | Stem cell biology | Research Institution |
| 87 | More visible commenting, including summaries and badges representing comments | As a reader, I want to | to read high-quality reviews on preprints, so that I can | better evaluate preprints | China | Stem cell biology | Research Institution |
| 88 | Dynamic layouts | As a reader, I want to | download all preprint figures as a powerpoint slide, so that I can | more easily incorporate them into journal clubs and other presentations | USA | Synthetic biology | Research Institution, Community bio |
| 89 | Browse data inline | As a reader, I want to | link out or browse genetic sequences inline, so that I can | better evaluate the preprint | USA | Synthetic biology | Research Institution, Community bio |
| 90 | Lay summaries of preprints (and searching for preprints with additional summary or review content) | As a (non-specialist) reader, I want to | read a lay summary and see a graphic abstract accompanying preprints, so that I can | better understand the preprint | USA | Synthetic biology | Research Institution, Community bio |
| 91 | Embed video, code, etc | As an author, I want to | include data and executable code and figures into my preprint, so that | my preprint can be interactive | USA | Synthetic biology | Research Institution, Community bio |
| 92 | Clearer documentation during submission | As an author, I want to | be prompted to share my reagents (eg plasmids through FreeGenes or Addgene under the OpenMTA or UBMTA) along with an explanation of the terms of the MTA (UBMTA vs OpenMTA), so that I can | make my science open | USA | Synthetic biology | Research Institution, Community bio |
| 93 | Embed video, code, etc | As an author, I want to | be able to embed SBOL files in my preprint, so that | others can reuse my synthetic biology parts | USA | Synthetic biology | Research Institution, Community bio |
| 94 | Helpdesk | As an author, I want to | speak to someone at my chosen preprint server prior to submission, so that I can | understand if my article will fit the scope | USA | Clinician / Neurobiology | Clinic, Research Institution |
| 95 | Better linking to journal versions | As a reader, I want to | see embedded links to published versions when a preprint cited in a bibliography is published, so that I can | better trust the foundation of evidence the article I’m reading is built on | USA | Clinician / Neurobiology | Clinic, Research Institution |
| 96 | Journal submission/review status | As a reader, I want to | see where the preprint I’m reading has been submitted (and possibly rejected), so that I can | gauge expert opinions of the preprint | USA | Clinician / Neurobiology | Clinic, Research Institution |



2 Comments