Project Overview
Amid the COVID-19 pandemic, preprints are being shared, reported on, and used to shape government policy, all at unprecedented rates and journalists are now regularly citing preprints in their pandemic coverage. As well as putting preprints squarely in the public eye as never before, presenting a unique opportunity to educate researchers and the public about their value, the rise in reporting of research posted as preprints has also brought into focus the question of how research is scrutinized and validated. Traditional journal peer review has its shortcomings and the number of ways research can be evaluated is expanding. This can be a problem for journalists and non-specialist readers who sometimes don’t fully understand the difference between preprints, peer-reviewed articles, and different forms of peer review. Media coverage can result in the sharing of information which may later not stand up to scientific scrutiny, leading to misunderstanding, misinformation and the risk of damaging the public perception of preprints and the scientific process.
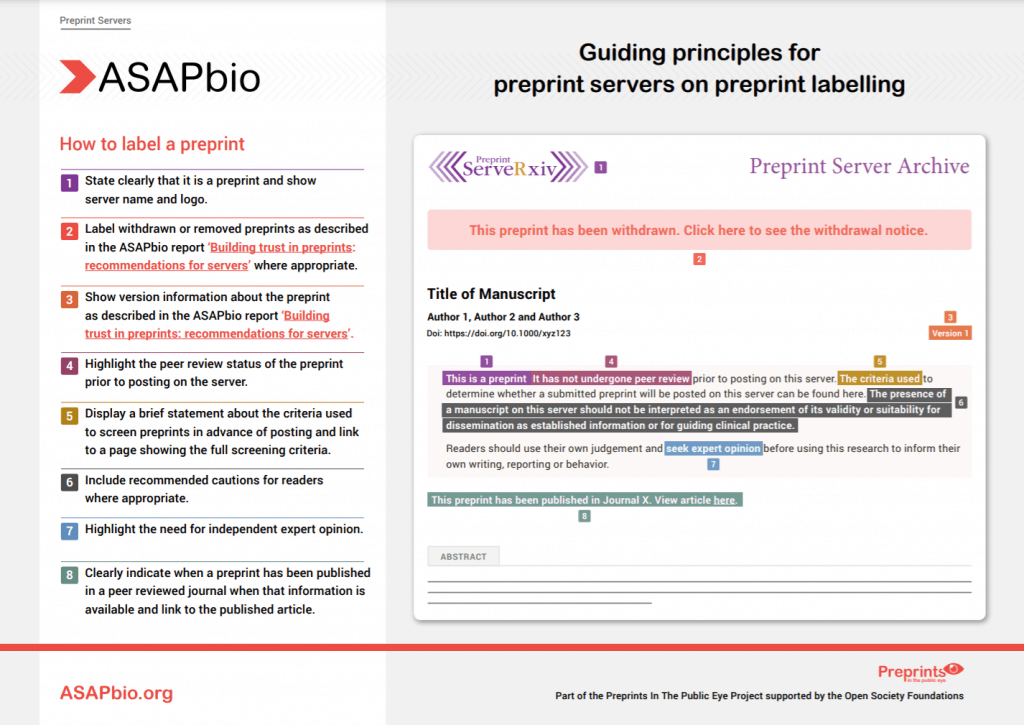
ASAPbio, with support from the Open Society Foundations, aimed to consolidate and expand on existing efforts to set best practice standards for reporting research posted as preprints via the launch of our Preprints in the Public Eye project. Read more in the project announcement. The outcome of this project is a set of four documents that set out guiding principles for the communication of research in the media for preprint servers, institutions, researchers, and journalists.
Outputs
Guiding principles infographics
Guiding principles full documents
News and updates
Project Participants
The following individuals contributed to the documents, but involvement does necessarily imply institutional endorsement.
- Dan Valen, Figshare
- Quincey Justman, Cell Systems, Cell Press
- Michael Markie, F1000 Research
- Theo Bloom, medRxiv
- Michele Avissar-Whiting, Research Square
- Alex Mendonça, SciELO
- James Brian Byrd, University of Michigan
- Shirley Decker-Lucke, SSRN
- Sowmya Swaminathan, Nature Research & Springer Nature
- Tom Ulrich, Broad Institute of MIT and Harvard
- Elisa Nelissen, KU Leuven
- Roberto Buccione, IRCCS Ospedale San Raffaele & Università Vita-Salute San Raffaele
- Emily Packer, eLife
- James Fraser, University of California San Francisco
- Kirsty Wallis, University College London
- Tanya Lewis, Scientific American
- Michael Magoulias, SSRN
Support for this project provided by the Open Society Foundations