ASAPbio is iteratively seeking community feedback on a draft model for a Central Preprint Service. We will integrate community and stakeholder feedback into a proposal, containing several model variants, to funders this fall. Please leave your feedback on utility of the Central Service, its features, and the model described in the Summary in the comment section at the bottom of the page, or email it privately to jessica.polka at gmail.com. More comments are posted on hypothes.is (follow this link and expand the menu at right)
Central Service model documents
- Summary: Background and a draft model
- Appendix 1: Rationale for a Central Service
- Appendix 2: Current feedback on Central Service features
Summary
At the ASAPbio Funders’ Workshop (May 24, 2016, NIH), representatives from 16 funding agencies requested that ASAPbio “develop a proposal describing the governance, infrastructure and standards desired for a preprint service that represents the views of the broadest number of stakeholders.” We are now holding a Technical Workshop to advise on the infrastructure and standards for a Central Service (CS) for preprints. ASAPbio will integrate the output of the meeting and community and stakeholder feedback into a proposal to funding agencies this fall. The funders may issue a formal RFA to which any interested parties could apply for funding. More details on this process are found at the end of Appendix 2.
Background
The preprint ecosystem in biology is already diverse; major players include bioRxiv, PeerJ Preprints, the q-bio section of arXiv, and others. In addition, platforms such as F1000Research and Wellcome Open Research are producing increasing volumes of pre-peer reviewed content. PLOS has a stated commitment to exploring posting of manuscripts before peer review, and other services may be developed in the future.
Increasing the number of intake mechanisms for the dissemination of pre-peer reviewed manuscripts has several advantages, for example: 1) generating more choices for scientists, 2) promoting innovative author services, and 3) increasing the overall volume of manuscripts, thus helping to establish a system of scientist-driven disclosure of their research. However, an increasing number of intake mechanisms also may lead to confusion and difficulty in finding preprints, heterogenous standards of ethical disclosure, duplication of effort in creation of infrastructure, and uncertainty of long-term preservation. (See a more complete discussion of why we think it is essential to aggregate content in Appendix 1.)
Based upon funder interest from the May 24th Workshop, ASAPbio will propose that funding agencies support the creation of a Central Service (CS) that will aggregate preprint content from multiple entities. This service will have features of PubMed (indexing/search) and PubMed Central (collection, storage, and output of manuscripts and other data).
The advantages of this system for the scientific community would be:
- Oversight by a Governance Body. The content, performances, and services of the CS would be overseen by a Governance Body composed of highly respected scientists and technical experts. The formation of Governance Body, which will have international representation and be transparent in its operation, will be addressed by a separate ASAPbio task force and will not be discussed in the Technical Workshop. The connection between the CS and a community-led Governance Body will ensure that preprints continue to serve the public good and develop in ways that benefit the scientific community, beyond the needs of individual publishers and servers. This formation of a central, well-functioning Governance Body has been repeatedly described by funders and scientists as an essential element in gaining respectability for preprints and guiding the system in the future.
- Guaranteed stable preservation. Archiving content through a CS better assures permanence of the scientific record, even if a preprint server/publisher decides to discontinue their services.This is a key feature for both scientists and funders.
- Greater discoverability and visibility for scientists. The CS would become the location for scientists to search for all new pre-peer reviewed content. Lessons from arXiv indicate that a highly visible, highly respected single site for searching for new findings is essential for the scientific community.
- Clarity on what qualifies as a respected preprint. Scientists want their preprint to “count” for hiring, promotions, and grant applications. However, universities and funding agencies are concerned about quality control for preprints and how they can guide their scientists and reviewers on what qualifies as a credible preprint or preprint server. The CS/Governance Body will work with universities and funders to apply uniform standards of author identity, checks for plagiarism, moderation of problems, and create ethical guidelines for research and disclosure. Thus, content on the CS, coming from several sources, will meet uniform guidelines acceptable to funders and universities.
- Better services for scientists. Scientists, as consumers, want better ways of viewing content. They want to read manuscripts in an xml format on the web or as a PDF download, more easily link to references, and more easily view figures and movies. The CS would perform document conversion to ease viewing and searching for material, thereby accelerating new discoveries. The CS would have an API to enable innovative reuse by other parties to provides services that could be valuable for scientists beyond the scope of the CS (e.g. evaluations of work, journal clubs, additional search engines).
- Reduced overall cost. The central service can efficiently provide services (such as archiving, automated screening, and document conversion) that otherwise would be provided redundantly by each intake server/publisher.
We discussed various models for the CS with stakeholders (see Appendix 2 for types of models and the feedback that we received). This document describes the current iteration of the model, which is still in draft form. We will present several variations to funders this fall, based on feedback received, including the comments here. If you prefer, you may email comments privately to jessica.polka at gmail.com.
The CS would undertake several functions including centralized document conversion, accrediting (via setting guidelines for intake), archiving, search, and an API for third-party use. We are currently considering that the CS would not display full-text, but instead would send back the converted full-text to the intake server for display.
In this draft model:
- Servers would facilitate the submission of a .doc or .tex file and a standardized set of metadata (e.g. authors names, potentially ORCID numbers, etc) to the CS. From this file, the CS could extract an html or xml file (possibly including links to references, figures, etc).
- If this file passes CS screening (including plagiarism detection, and potentially human moderation etc), it would be admitted into the central database, assigned a unique ID, and be sent back to the intake provider for display.
- The CS would archive the original .doc file and other associated files, and also make these available via an API; as reference extraction technology improves, etc, new html/xml derivatives can be prepared. The CS would reserve the right to display content if the intake provider is not able to do so or if required by the funders or governance body.
- Readers could search for preprints (or receive alerts) through CS-hosted tools that would display metadata (including abstracts); readers would be sent to the intake server for full-text display of preprints.
- All aspects of the central service would be under the control of a governing body, which would have international representation from the scientific community and could develop over time.
The Technical Workshop will discuss the features, mechanisms, existing infrastructure, potential concerns and challenges, and timelines for implementation for the elements in orange on the diagram below.
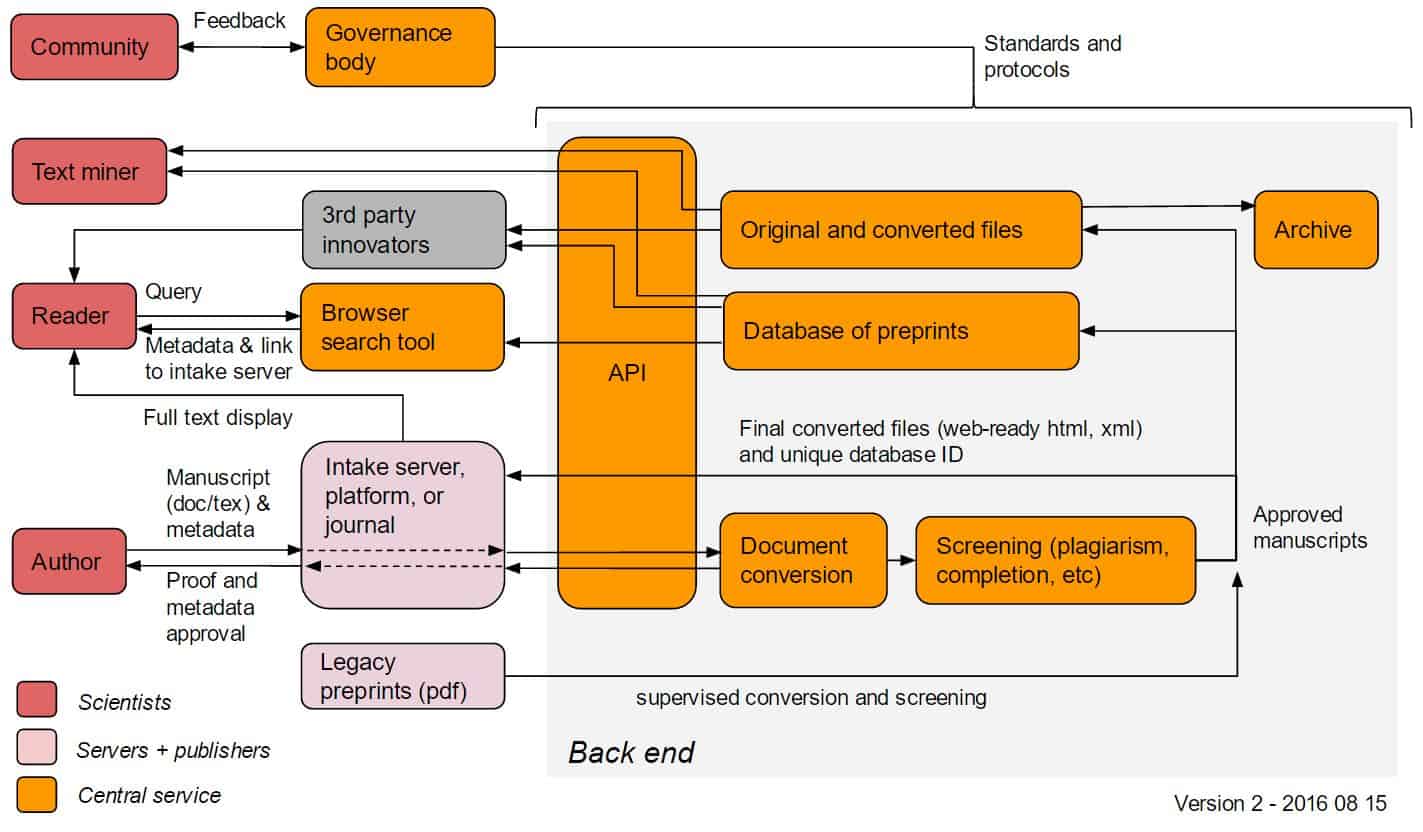
ASAPbio will continue to modify the model before and after the Technical Workshop before presenting several variations to funders in the fall.
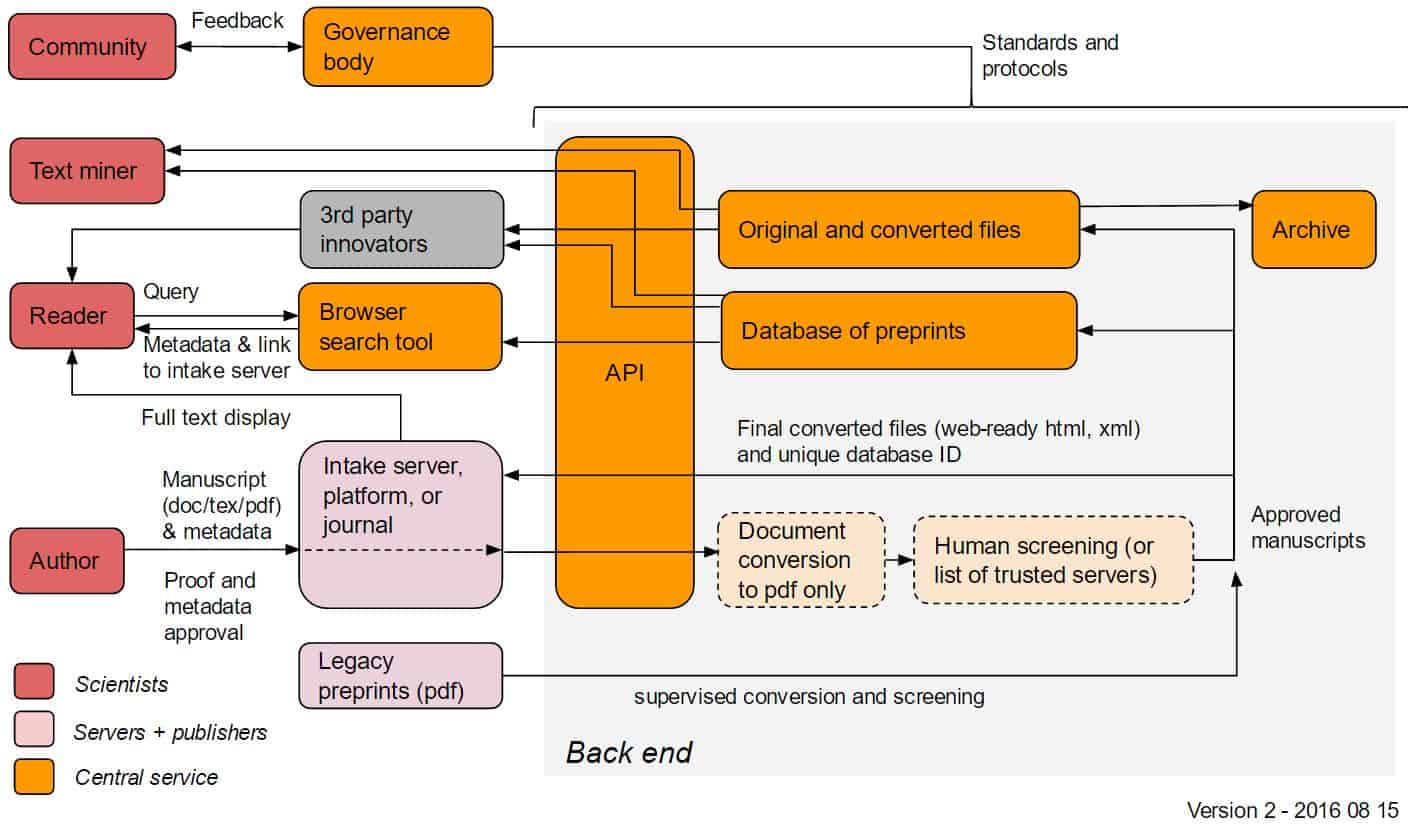
Below: possible early-stage implementation


Hi Jessica, I really love the idea of a centralized preprint server, but not quite yet. I still feel like the best model needs to be worked out. For instance, I really like that PeerJ offers me the opportunity to post protocols, when biorXiv doesn’t, but in general I much prefer the simplicity of biorXiv.
Additionally, many of the problems, like quality-control, trust and anti-trolling still really need innovative solutions.
Having said that, I feel there is definitely room for, and a need for, some sort of aggregator and repository that combines and stores feeds from all preprint servers.
I feel a niche that this could really serve is to offer an ability to popularize a preprint, to get lots of eyes on it, and disseminate comments. For instance, in the case of the results of a clinical trial that Les Loew brought up in his editorial on preprints in Biophysical Journal, I think you could really strengthen peer review by considering comments.
–Buz
Thanks for the feedback, Buz! I agree with you – I don’t think a central *server* is the right solution, but I believe a central *service* could be useful. Especially one that provides access to all preprints via an API; this would enable tool-builders to construct services like the commenting platform you’re describing.
This is a great idea! I think the service model could enable even more innovation rather than diminish it, as it could give visibility to smaller, upstart preprint servers.
It should work just like PubMed and PubMedCentral – index abstracts and store pdf copies provided by pre-print servers. And possibly provide possibility to comment, but this would require constant moderation efforts.
I think the CS idea is great (shame it didn’t get built in the 90s!). My only technical nitpick is that it shouldn’t require everything in TeX or .doc format. I think that’ll end up being quite restrictive in the not-so-long term.
There isn’t a simple answer to that question, but being open-ended here is better than not.
Mandating formats might not even be necessary, if the CS is not going to serve up full manuscripts. Mucking around with format conversions and so on seems like a different job from the role of centralized archive.
Exactly. It should work just like PubMed and PubMedCentral – index abstracts and store pdf copies provided by pre-print servers.
I like the diagram of the technical architecture. Some suggestions:
It is a good idea to have the early stage implementation so that automatic document conversion other than to PDF isn’t part of the first implementation. This is a complex topic and goes beyond what preprint servers currently offer, so I can see document conversion to HTML and XML coming later. The same goes for other input formats beyond .docx and .tex. But I would not allow .pdf as input in the early stage implementation, as this makes it harder to convert to other formats later.
The early stage implementation also needs Document Conversion (limited to .docx and .tex input and .pdf output) and Screening. The difference to later implementations could be that the screening process is simpler, e.g. manual approval by a small group of editors rather than a semi-automated workflow.
The index (I assume using Solr or Elasticsearch) should be part of the backend, and the API should be built on top of the index (not the database). All frontend services including search should use the API, rather than talk to the index or database directly.
File storage is an important part of the backend and would therefore add it as a box in the diagram. It might for example not be a good idea to store the fulltext of all documents directly in the database, but rather store links to them hosted in some file system or cloud bucket. The search index will probably also have the fulltext, but maybe not in the early stage implementation.
Thank you for these extremely helpful comments, Martin. We’ve updated the diagrams to reflect your suggested changes (originals are linked below the new versions).
Its seems to me, once you figure out what fields are desired in the database (full text please!) this could be brought online very quickly. You can get a full, scalable, ElasticSearch database up and going on Amazon Cloud in less than a day… And that comes with a web interface for searches, and easily managed permissions as to who can write to / read from the database.
Just curious, why keep the front end going though an API, when it is already built into elasticsearch? If the hosting is scalable per the load on the cloud, why not?
Will it be possible for users to build APIs on top of the central server? It would be an essential feature.
Thanks Stefano – I agree that allowing free access to the data (for both humans and machines) is extremely important. A uniform license (like CC-BY) for preprints in the database would also be important for promoting innovative reuse.
I’ve followed this discussion including Casey Greene’s inline comments. While I do think we need to be careful not to just create another journal, there are some big problems with preprints that I don’t how we can solve without a CS.
For example, we want preprints to be recognized as part of the literature and therefore citable. However, what happens when a person submits their preprint to two, three, or more preprint servers. Which preprint do we cite? And if we want to start a discussion on the paper which server do we comment on?
We can’t rely upon the preprint servers themselves to prevent dual publication. Even when a server like PeerJ Preprints as the policy: “PeerJ Preprints does NOT accept submissions which have been identically posted on other preprint servers” how do they enforce it?
A person could just first submit their preprint to PeerJ, then submit it to other preprint servers with relaxed policies. And this is currently happening. I know which articles have been cross posted at multiple servers, although it is a minority of preprints at the moment.
And if we want preprints to be taken seriously I do think we need to be a little bit more strict about what gets published as a preprint. For example, there are literally preprints that I have seen in my database which are only an abstract, that’s it. Maybe a simple minimum word count would suffice?
Having a central place where comments are moderated/stored would also be really nice. I find that people submitting preprints are open to feedback and welcome comments, even if they might be critical. Just as there are “superusers” on Quora who have developed a reputation for the quality of their answers, I could see researchers who constantly provide insightful feedback on preprints as gaining recognition. And maybe some of these comments could even be seen as a “review” of the article or given a DOI.
I am unsure about the technical aspects of the CS, but if you need information from PrePubMed let me know.
The risk is that you make something centralised and vital and then the funding runs out. Right from the get-go you should have an exit strategy of what happens if this service can’t be supported anymore. Obviously you should accept formats that your system doesn’t understand (yet) as you’ll otherwise be a drag factor to innovation and the posting of data with papers. The service should be distributed in such a way that it can survive the collapse of the core funding.
Agree that long-term funding business plan is the single most important issue.
That’s not what I said. The most important issue is ensuring that it’s not fragile and can survive beyond the initial model.
I completely agree that centralization of some kind is critically important for the ASAPbio effort. Whatever that centralization looks like, it needs to be flexible enough to accommodate functionality of the future: new formats, new search, new index, new review, new social media. My greatest fear is that we will have an explosion of pre-print servers launched by every commercial and society journal, and many new independent startups (this is already happening). In time, many of these will sputter out. Therefore any CS aggregator needs to be structured for long-term financial stability.
I still strongly favor the arXiv model with 1 pre-print server for biology that is its own CS, that is stably funded by some common fund of users, government, and private philanthropy, and that is a not-for-profit entity. The ASAPbio community will have more control over the governance of this server, how the content is served up and quality-controlled, how and if there is commenting and PPPR, etc. The only service out there that meets these criteria at the moment is bioRxiv. I think we as a community need to settle on a single CS (bioRxiv or a new entity as imagined above) in the expectation that all or most of the other new independent pre-print servers will fail in a few years.
Forgot the important disclosure that I am a member of the bioRxiv Advisory Board.
A big worry I have about centralization is that if the central server is doing all the screening and conversion and the budget gets tight, there will be pressure to cut the scope. So figuring out how willing the funders are to commit to scale for the long term would be important.
I am not well versed in the technical side, but it seems to me that word as an input format is a really bad idea. Doesn’t that mean that figures will be of poor quality?
Ivan, great points about the long-term plan. Re images, the system would absolutely need a way to include attachments (like supplementary files, etc) and some of these could of course be figures. Conversion software like Aperta (which is already operational for PLOS Biology submissions) can automatically place figures back into text extracted from a word file.
My $0.02.
I do like the idea of a CS that will aggregate content from the growing (which is positive in itself) number of preprint entities – https://en.wikipedia.org/wiki/Preprint
I did watch the talks from the #ASAPbio meeting back in Feb. and some of them again over the last 2 days.
As a strong supporter of OA, I’m clearly all for preprints. I’ve co-authored two life sciences ones myself and I’m not even a researcher _per se_.
My main concern at present with regards to preprints is *the lack of uptake generally* compared to the success of arXiv/physics & maths etc. About 8,000 preprints a month.
IMO, the situation is not that different with regards to Institutional Repositories (IR) and green OA. Lot’s of IRs but uptake is generally low and generally, only about 15% of content is OA.
Unless mandated to do so, researchers (unfortunately) generally tend not to be concerned about whether or not their papers are made OA either through the green or gold routes. Other than the physics/maths field, researchers tend not to embrace using preprints.
All in all, at this stage, I’m less concerned about whether or not their is a CS or not, but more about things that can be done to encourage/reward researchers to embrace preprints !
My $0.02.
I do like the idea of a CS that will aggregate
content from the growing (which is positive in itself) number of
preprint entities – (See the Wikipedia preprint page).
I did watch the talks from the #ASAPbio meeting back in Feb. and some of them again over the last 2 days.
As
a strong supporter of OA, I’m clearly all for preprints. I’ve
co-authored three life sciences ones myself and I’m not even a
researcher _per se_.
My main concern over the years and still with
regards to preprints is *the lack of uptake* generally compared to the
success of arXiv/physics & maths etc. About 8,000 preprints a month.
IMO,
the situation is not that different with regards to Institutional
Repositories (IR) and green OA. Lot’s of IRs but uptake is generally low
plus only about 15% of content is OA.
Unless mandated to do so,
researchers (unfortunately) generally tend not to be concerned about
whether or not their papers are made OA either through the green or gold
routes. Other than the physics/maths field, researchers tend not to
embrace using preprints.
All in all, at this stage, I’m less
concerned about whether or not their is a CS, but more about what can be
done to encourage/reward researchers to post preprints !